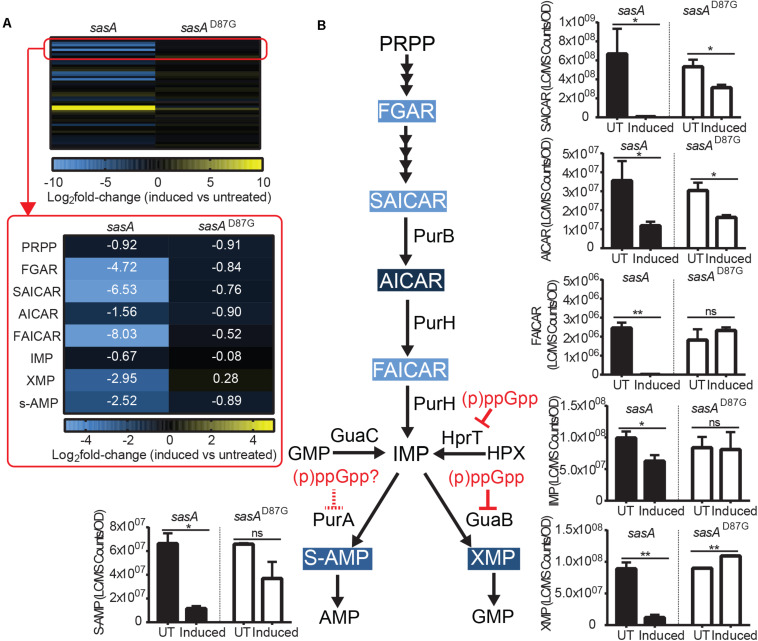
FIGURE 5.
Purine precursors are depleted during SasA expression. (A) Heat map of metabolite changes in cells after sasA or sasAD87G expression. Red box highlights the changes in the level of PRPP-IMP pathway metabolites in addition to adenylosuccinate (S-AMP) and xanthosine monophosphate (XMP). PRPP, Phosphoribosyl pyrophosphate; FGAR, Phosphoribosyl-N-formylglycineamide; SAICAR, Phosphoribosylaminoimidazolesuccinocarboxamide; AICAR, 5-Aminoimidazole-4-carboxamide ribonucleotide; FAICAR, 5-Formamidoimidazole-4-carboxamide ribotide; IMP, Inosine monophosphate. Numbers indicate mean fold-change in binary logarithm relative to untreated cells. (B) Summary of metabolite changes in the de novo purine biosynthesis pathway in SasA-expressing cells. Bar plots shown are levels of SAICAR, AICAR, FAICAR, IMP, S-AMP, and XMP before and after sasA or sasAD87G expression. PurB, Adenylsuccinate lyase; PurH, Phosphoribosylaminoimidazole carboxamide formyltransferaseand inosine-monophosphate cyclohydrolase; GuaC, GMP reductase; HprT, Hypoxanthine phosphoribosyltransferase; PurA, Adenylosuccinate synthetase; GuaB, IMP dehydrogenase. UT: untreated, Induced: after induction. Data shown are LC-MS ion counts normalized to OD600. Error bars indicate SD. n = 2. **p < 0.01, *p < 0.05, ns: not significant (Student’s t-test). Color-shaded are metabolites with log2 fold-change > 2 as shown in (A). Solid and dashed red arrows indicate known and speculated inhibition by (p)ppGpp respectively.