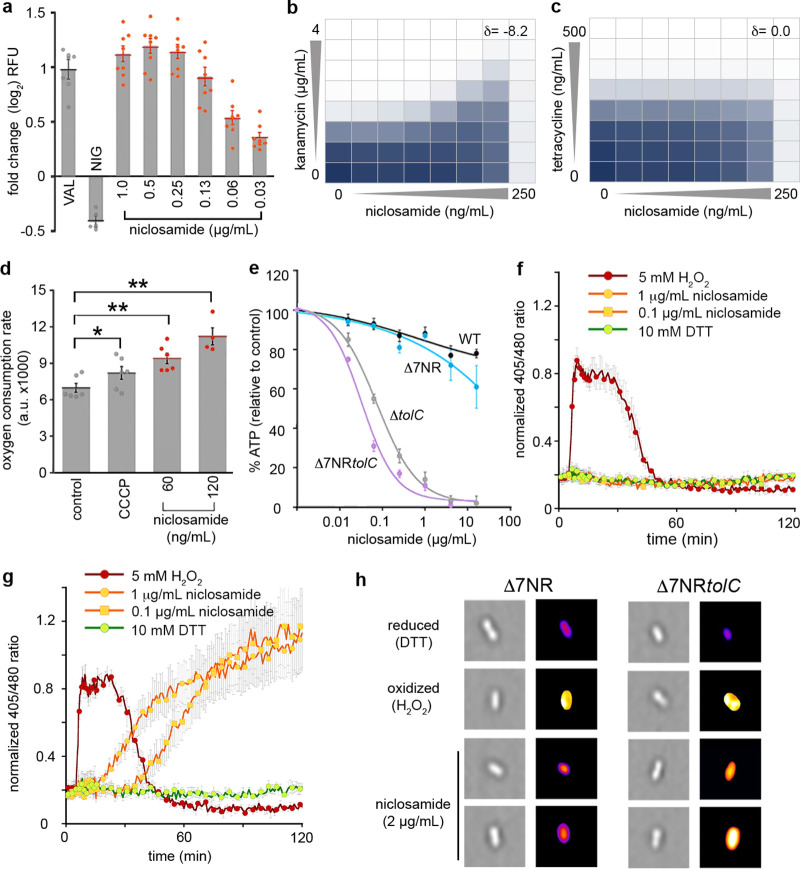
FIG 2.
Antibiotic mechanisms of niclosamide. (a) Fold change in DiSC3(5) fluorescence. E. coli was grown in MHB with 10 mM EDTA to an OD600 of 1. Cells were incubated with DiSC3(5) for 20 min prior to administration of 0.5 μg/ml valinomycin (VAL; a ΔΨ-dissipating ionophore), 0.5 μg/ml nigericin (NIG; a ΔpH-dissipating ionophore), or 0.03 to 1 μg/ml niclosamide. KCl (100 mM) was added to cells prior to valinomycin treatment. RFU, relative fluorescence units. (b and c) The combined inhibitory effects of 0 to 250 ng · ml−1 niclosamide and either (b) 0 to 4 μg · ml−1 kanamycin or (c) 0 to 500 ng · ml−1 tetracycline were tested against the Δ7NRtolC strain in a checkerboard format. Bacterial growth is shown as a heat plot. (d) Oxygen consumption was measured using the MitoXpress oxygen probe in Δ7NRtolC cells (mid-log phase; OD600 = 0.15) overlaid with mineral oil for 20 min. a.u., arbitrary units. (e) Relative cellular ATP levels were estimated by luciferase activity and compared to an unchallenged (DMSO-only) control. (f and g) Intracellular oxidation levels were measured in (f) WT E. coli and (g) ΔtolC strains constitutively expressing redox-sensitive GFP (roGFP) following administration of 5 mM H2O2 (oxidized control), 10 mM DTT (reduced control), or niclosamide. (h) Representative high-throughput fluorescence microscopy images of Δ7NR and Δ7NRtolC cells 120 min after administration of DTT, H2O2, or niclosamide. Images on the right are pseudocolored ratio images after analysis with ImageJ. Panels a to g were constructed from pooled data from at least three independent biological replicates. Labels indicate significant responses over the control (* = P < 0.05; ** = P < 0.01). Statistical analyses were performed using one-way analysis of variance (ANOVA) and the Kruskal-Wallis test. Error bars indicate SEM.