Figure 2.
Insertion of a Stem-Loop Sequence before the Poly(A) Signal Attenuates Cell Transformation Induced by Polyadenylation of H3.1 mRNA
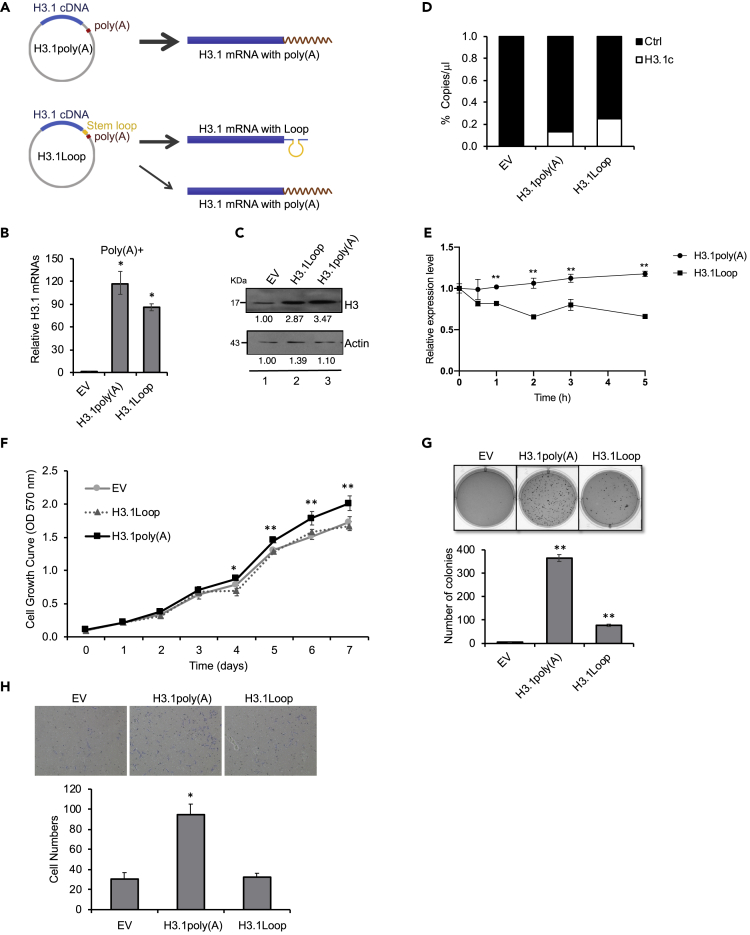
(A) An H3.1 cDNA fragment was inserted into the MCSs of pcDNA vector, which contains a poly(A) signal in the downstream of MCSs. H3.1Loop plasmid: a stem-loop sequence was inserted before the poly(A) signal, whose transcription terminates near the stem-loop sequence, generating H3.1 mRNAs mostly without poly(A) tail. pcDNA-Empty (EV), H3.1poly(A), or H3.1Loop vector was stably transfected into BEAS-2B cells.
(B) RT-qPCR results using oligo (dT) primers for reverse transcription to capture polyadenylated H3.1 mRNAs. The data shown are the mean ± S.D. (n = 3). Student's t test was applied for statistical significance: ∗p < 0.01.
(C) Western blot analysis. The band intensities were quantified using ImageJ software. The controls in lane 1 were used as references (set to 1).
(D) Digital droplet PCR (ddPCR). Total DNA was extracted from BEAS-2B cells stably expressing H3.1poly(A) or H3.1Loop plasmid. The copy number of exogenous H3.1 cDNA was analyzed by ddPCR with primers designed to amplify a region spanning T7 and H3.1 cDNA on the vector. The ribonuclease P/MRP 30 kDa subunit (RPP30) was used as an internal control. The data shown are the mean ± S.D. (n = 3).
(E) RNA stability assay for exogenous H3.1poly(A) mRNA and H3.1Loop mRNA. BEAS-2B cells stably expressing H3.1poly(A) or H3.1Loop were treated with 10 μg/mL actinomycin D for the indicated time. Total RNA was extracted, and the mRNA level was analyzed by qPCR with primers designed to amplify the exogenous H3.1 mRNA. The relative expression level was normalized by the reference gene GAPDH. The data shown are the mean ± S.D. (n = 3). Student's t test was applied for statistical significance: ∗∗p < 0.01.
(F) The cell growth rate was measured by MTT. The data shown are the mean ± S.D. (n = 3). Student's t test was applied for statistical significance: ∗p < 0.05; ∗∗p < 0.01.
(G) Soft agar assays. The data shown are the mean ± S.D. (n = 3). Student's t test was applied for statistical significance: ∗∗p < 0.01. See also Figure S1.
(H) Transwell invasion assays. The migrated cells were fixed, stained, and counted. Cell number was determined and averaged from three fields randomly selected from each sample. Bars indicate S.D. (n = 3). Student's t test was applied for statistical significance: ∗p < 0.05.