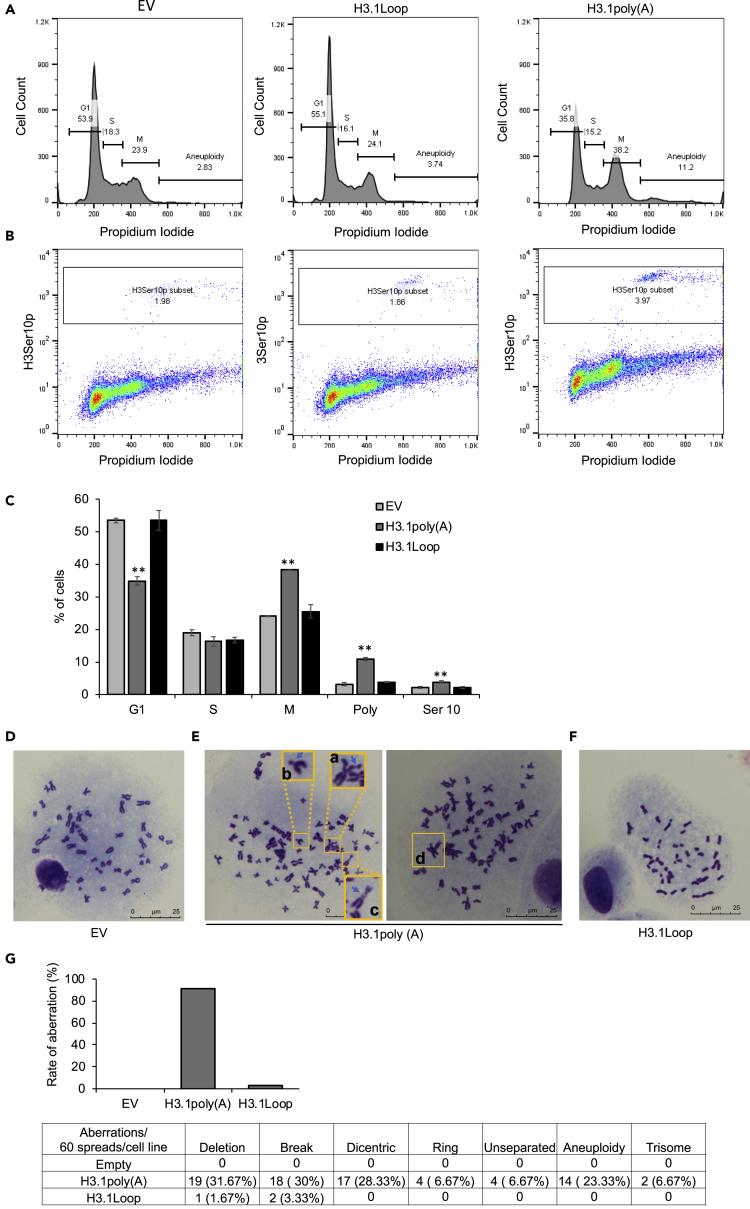
Figure 5.
Polyadenylation of H3.1 mRNA Causes Mitotic Arrest and Genomic Instability
(A) Flow cytometry analysis of the cell cycle. BEAS-2B cells that transiently transfected with empty vector (EV), H3.1Loop, or H3.1poly(A) were stained with PI.
(B) Flow cytometry analysis of the cells stained with antibodies specially against phosphorylated H3S10 (H3S10p) to monitor M phase cells.
(C) The experiment shown in (A) and (B) was repeated three times, and statistical analysis on the data was performed. The plot was represented as the percentage of cells in each cell cycle phase, polyploid/aneuploid cells, or H3S10p positive cells. The data shown are the mean ± S.D. from experiments performed in triplicate. Student's t test was applied for statistical significance: ∗∗p < 0.01.
(D–G) Giemsa-stained spread chromosomes from BEAS-2B cells transfected with EV, H3.1poly(A), or H3.1Loop plasmid. (D) Metaphase spread from control cells showing normal chromosome spread (scale bar, 5 μm). (E) Metaphase spread from H3.1poly(A) cells showing abnormal chromosome spread (scale bar, 5 μm), including chromatid break (a), deletion (b), dicentric chromosome (c), triradial chromosome (d) etc. (F) Metaphase spread from H3.1Loop cells showing mostly normal chromosome spread (scale bar, 5 μm). (G) Quantification of frequency of chromosome aberration after BEAS-2B cells were transfected with EV, H3.1poly(A), or H3.1Loop plasmid. The plot was represented as the rate of aberrations per 60 chromosome spreads for each cell line. The table was represented as the ratio of different types of aberration per 60 chromosomes for each cell line.