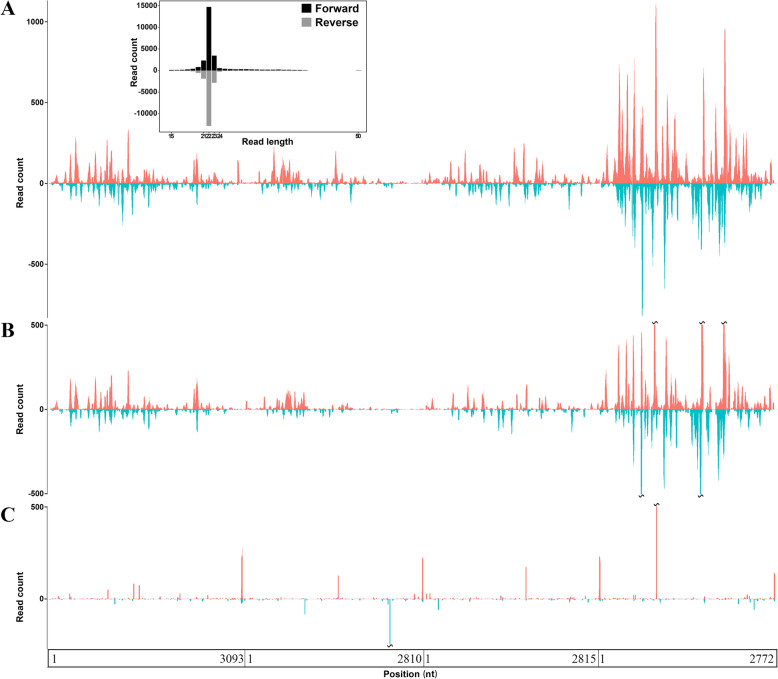
Fig. 1.
Genome coverage and average depth of the MGTV strain Yunnan2016. The y-axis represents the read-counts for each genomic position. a. The x-axis represents positions in the reference genome of the MGTV strain Yunnan2016 (GenBank: MT080097, MT080098, MT080099 and MT080100) and the sRNA-seq data SRR4116826 (Table 1) was aligned to this reference genome; b. The x-axis represents positions on the reference genome of the MGTV strain Xinjiang2016 (GenBank: MK174251, MK174244, MK174230 and MK174237) and the sRNA-seq data SRR4116826 (Table 1) was aligned to this reference genome; c. The x-axis represents positions on the reference genome of the MGTV strain Yunnan2016 and the sRNA-seq data SRR8439389, SRR8439390, SRR8432408, SRR8432409, SRR811197093 and SRR811197094 (Table 1) were aligned to this reference genome as negative controls