Graphical abstract
Keywords: Calcium coordination, Bacterial adhesin, Adhesion protein, Beta-sandwich domains
Abbreviations: MhLap, Marinobacter hydrocarbonoclasticus long adhesion protein; AhLap, Aeromonas hydrophila long adhesion protein; MpIBP, Marinomonas primoryensis ice-binding protein; RTX, Repeats in toxin; vWFA, von Willebrand Factor A
Highlights
-
•
Elongated beta-sandwich repeats are a major part of bacterial RTX adhesins.
-
•
The repeats are arranged in tandem to extend away from the bacterial surface.
-
•
Calcium ions are coordinated in the linkers between repeats to stiffen the protein.
-
•
Rigidification of the tandem repeats further helps extension of the adhesin.
-
•
The repeats differ greatly between species, but all have Ca2+ in their linkers.
Abstract
RTX adhesins are long, multi-domain proteins present on the outer membrane of many Gram-negative bacteria. From this vantage point, adhesins use their distal ligand-binding domains for surface attachment leading to biofilm formation. To expand the reach of the ligand-binding domains, RTX adhesins maintain a central extender region of multiple tandem repeats, which makes up most of the proteins’ large molecular weight. Alignments of the 10-15-kDa extender domains show low sequence identity between adhesins. Here we have produced and structurally characterized protein constructs of four tandem repeats (tetra-tandemers) from two different RTX adhesins. In comparing the tetra-tandemers to each other and already solved structures from Marinomonas primoryensis and Salmonella enterica, the extender domains fold as diverse beta-sandwich structures with widely differing calcium contents. However, all the tetra-tandemers have at least one calcium ion coordinated in the linker region between beta-sandwich domains whose role appears to be the rigidification of the extender region to help the adhesin extend its reach.
1. Introduction:
The beta sandwich is a ubiquitous protein fold. In its most basic form, the domain consists of at least two antiparallel beta sheets, the amphipathic nature of which leads to a hydrophobic core that holds the secondary-structure elements together (Chothia and Janin, 1981, Cohen et al., 1981). While additional strands or alpha helices are often present, these elements are not a prerequisite for all beta sandwiches. Furthermore, the number of strands that make up each beta sheet can differ between families (Chothia et al., 1997), leading to a diverse collection of domains that – though all part of the sandwich-like proteins group – maintain a variety of shapes and sizes.
There are at least 69 protein superfamilies that fall under the sandwich-like protein umbrella (Kister et al., 2002), with many of the best known being named after the heavily studied eukaryotic forerunners of the group. These include immunoglobulin-like, fibronectin III-like, and cadherin-like families (Bork et al., 1994, Leahy et al., 1992, Shapiro et al., 1995). Interestingly, the titular representatives for all these families are extracellular proteins that take part in ligand recognition and/or cellular adhesion (Singh et al., 2010, Takeichi, 1990, Williams and Barclay, 1988). Indeed, beta-sandwich domains appear well suited to these types of protein, being incorporated into extracellular proteins in both eukaryotes and prokaryotes with remarkable frequency. For example, regions of tandem beta-sandwich repeats are a hallmark of many adhesion proteins: a group of large, repetitive proteins that facilitate contacts between cells and their environment (Klemm and Schembri, 2000). These regions are found in both multi-cellular organisms (ex. integrin and cadherin, which maintain contacts between cells and extracellular matrix components (Campbell and Humphries, 2011, Takeichi, 1990)), and in single-celled organisms, where adhesion proteins allow bacteria to stick to surfaces (Guo et al., 2012, Ivanov et al., 2012, Leo et al., 2010, Schilling et al., 2001, Wagner et al., 2014) and cluster together to make bacterial communities known as biofilms (Borlee et al., 2010, Dong et al., 2020, Kikuchi et al., 2005, Martínez-Gil et al., 2010).
In all cases, the purpose of the tandem beta-sandwich domains appears to be the provision of an extended reach that projects the ligand-recognizing domains of the protein away from the cell surface (Gerlach et al., 2008, Guo et al., 2012). The beta-sandwich fold is well adapted to this role. The domain’s N and C termini are oriented opposite of each other, allowing tandem repeats to proceed in a linear fashion. Beta-sandwich domains are often exceptionally strong, maintaining their fold under intense strain; beta sandwiches from adhesion proteins, cohesins, and the muscle protein titin are able to remain folded when subjected to forces above 300 pN (Li et al., 2002, Lu et al., 1998, Oude Vrielink et al., 2017, Valbuena et al., 2009). Additionally, many beta sandwiches augment their fold or stability through the coordination of divalent cations, which are rare within but abundant outside the cell. Cadherins are known to bind calcium in the otherwise flexible loop regions between beta-sandwich domains, thereby rigidifying the structure and facilitating the homotypic interactions between cadherin molecules (Boggon et al., 2002, Kim et al., 2011, Koch et al., 1997). A similar calcium-dependent strategy has been recently discovered in bacterial adhesion proteins (adhesins). The extender beta sandwiches from an epithelial adhesin, SiiE, produced by Salmonella enterica require calcium for thermal stability and resistance to proteolysis (Peters et al., 2017), while the massive ice-binding adhesion protein from the Antarctic bacterium Marinomonas primoryensis completely loses its tertiary structure in the absence of calcium (Guo et al., 2013). Like cadherin, structure determination of small segments from both proteins showed the presence of calcium ions that coordinate to the linkers between beta-sandwich domains (Griessl et al., 2013, Vance et al., 2014), thereby producing a rod-like structure crucial for proper adhesion.
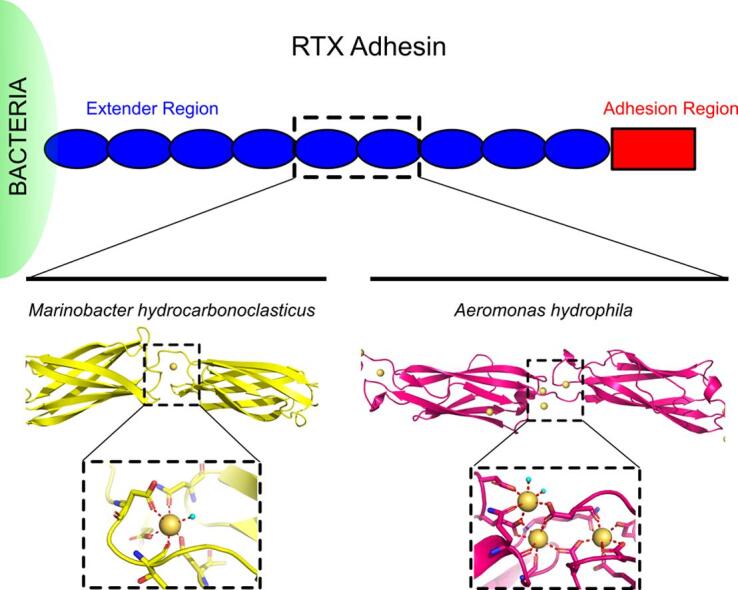
SiiE and the M. primoryensis ice-binding protein (MpIBP) belong to the same family of adhesion proteins, known as the RTX adhesins, which are widespread amongst Gram-negative bacteria (Guo et al., 2019a, Guo et al., 2019b, Satchell, 2011). RTX adhesins have been implicated in a variety of bacterial survival strategies, including pathogen infection (Cirillo et al., 2001, Syed et al., 2009, Wagner et al., 2014), symbiotic colonization (Hinsa et al., 2003, Martínez-Gil et al., 2010), and microbial community development (Guo et al., 2017). All known examples maintain some number of beta sandwiches for extension, though the exact number of repeats, protein family in which they reside, and sequence identity vary drastically between proteins (Guo et al., 2019a, Guo et al., 2019b). While it is assumed that all these regions require calcium, only the two aforementioned examples have been structurally characterized.
In this study, we analyzed extender regions from two additional RTX adhesins, including the long adhesion proteins from Marinobacter hydrocarbonoclasticus (MhLap) – an oil-eating marine bacterium – and Aeromonas hydrophila (AhLap) – an opportunistic pathogen of fish and mammals. From these proteins, constructs comprised of four beta-sandwich repeats, called tetra-tandemers, were produced and subjected to structural determination and biophysical characterization. Here we show that while the sequences, structures, and calcium requirements of these domains differ widely, the strategy of using calcium for rigidification of the linker between domains is consistent.
2. Materials and methods
2.1. Molecular cloning, protein expression and purification
To produce the tetra-tandemers for MhLap (NCBI: WP_014422746) and AhLap (NCBI: WP_011707240), genes encoding four of the beta-sandwich repeats in tandem were synthesized by GeneArt (Thermofisher). As previously described, codon optimization for expression in Escherichia coli was balanced with differentiating the repeats using codon degeneracy (Vance et al., 2014). These genes (Fig. S1) were then ligated into pET28a vectors using NdeI and XhoI restriction cut sites, thereby encoding an N-terminal His6-tag, and transformed in Top10 competent E. coli cells (Thermofisher) for plasmid amplification. Clones containing tetra-tandemer inserts were confirmed by Sanger sequencing (Robart’s Sequencing Facility, London, Ontario, Canada) and their plasmids electroporated into BL21(DE3) E. coli cells (Thermofisher) for expression.
Protein production was conducted as previously described, with minor modifications (Vance et al., 2014). Briefly, 1 L of LB broth was inoculated with an overnight 25-mL culture in the presence of 0.1 mg/mL kanamycin, followed by incubation at 37 °C with shaking. At an OD600 of 0.6, the 1-L culture was transferred to 23 °C and allowed to reach an OD600 of 0.9 before induction with 1.0 mM isopropyl β-D-thiogalactose (IPTG). Following overnight expression, cells from the culture were pelleted and resuspended in 50 mM Tris-HCl (pH 9), 500 mM NaCl, and 2 mM CaCl2. The cells were lysed by sonication, and the cell debris pelleted by centrifugation.
Purification of the tetra-tandemers required a two-step process. 1) Nickel-affinity chromatography: the proteins were incubated with nickel NTA agarose resin (Qiagen) and washed with resuspension buffer + 5 mM imidazole. After 3 column volumes of washing, the protein was eluted using resuspension buffer + 400 mM imidazole. 2) Size-exclusion chromatography: fractions from the nickel-affinity chromatography were pooled and concentrated to 5 mL for injection onto a Supderdex-200 16/60 column (GE Healthcare). Running buffer contained 50 mM Tris-HCl (pH 9), 200 mM NaCl, and 2 mM CaCl2. Purity of fractions containing tetra-tandemer was tested using 10% SDS-PAGE.
2.2. Calcium titration via circular dichroism spectroscopy
Calcium ions were removed from the protein through dialysis in 10 mM Tris-HCl (pH 9.0), 50 mM NaCl, and 5 mM EDTA. After several changes in this high EDTA buffer, the protein was dialyzed in a low-EDTA version of the same buffer (0.01 mM EDTA) and diluted to 15 µM in this buffer for circular dichroism spectroscopy. Small volumes (1 µL) of concentrated CaCl2 (92.5 mM) were added directly to the 200-µL sample in the cuvette to give incremental increases of 0.5 mM Ca2+. Twelve scans were taken at 23 °C using a Chirascan CD Spectrometer (Applied Photophysics) for each addition. The scans were averaged, buffer reference-subtracted, and subjected to three-point smoothing using PROVIEWER software. Deconvolution was performed with OLIS SpectralWorks (On-Line Instruments).
2.3. Crystallization and structure determination
For crystallography trials, the tetra-tandemers were buffer exchanged into 20 mM Tris-HCl (pH 9) and 10 mM CaCl2 and concentrated to 15 mg/mL (MhLap tetra-tandemer) or 18 mg/mL (AhLap tetra-tandemer). Microbatch methods were used in which 1 µL of protein was added to 1 µL of different precipitant solutions and covered with 11 µL of paraffin oil. Following optimization from initial crystal hits, both constructs formed well-diffracting, needle-like crystals, but under different conditions. MhLap tetra-tandemer crystallized in 0.1 M sodium acetate (pH 5.0), 0.5 M sodium chloride, and 20% PEG 6000; AhLap tetra-tandemer crystallized in 0.1 M sodium acetate (pH 4.6), and 9 – 13% PEG 20, 000.
Structures for both MhLap and AhLap tetra-tandemers were solved using calcium phasing. For the MhLap tetra-tandemer, the single-wavelength anomalous diffraction datasets were collected at a wavelength of 1.77121 Å on CLSID08-1 beamline at the Canadian Light Source (CLS). For the AhLap tetra-tandemer, a single-wavelength anomalous diffraction dataset was collected on a home X-ray source diffractometer equipped with a chromium rotating anode producing X-rays at a wavelength of 2.2909 Å; a subsequent high-resolution native dataset was collected on CLSID08-1 beamline at the CLS. The datasets were indexed and integrated using XDS (Kabsch, 2010) and scaled and merged by AIMLESS in CCP4 suite (Evans, 2006, Winn et al., 2011). The Ca2+-SAD datasets for MhLap and AhLap tetra-tandemers were run through Phenix-AutoSol (Liebschner et al., 2019, Terwilliger et al., 2009). The initial phases were calculated at 2.5 Å resolution for both proteins, and then extended to 2.0 Å for the MhLap tetra-tandemer, followed by Phenix-Autobuild (Terwilliger et al., 2008), BUCCANEER autobuild in CCP4 suite (Cowtan, 2006) and manual model-building using COOT (Emsley et al., 2010). At this point, the solved low-resolution model of the AhLap tetra-tandemer was used as a search model for a molecular replacement solution of the high-resolution data, using Phenix Phaser (McCoy et al., 2007). Both high-resolution structures were refined with Phenix-Refine (Afonine et al., 2012) and Refmac5 (Vagin et al., 2004, Winn et al., 2011). Crystallographic data collection and refinement statistics are summarized in Table 1. Tetra-tandemer structures were submitted to the Protein Data Bank with PDB codes 6XI3 (MhLap) and 6XI1 (AhLap).
Table 1.
Structural parameters for MhLap and AhLap tetra-tandemers.
| Protein sample | MhLap | AhLap | AhLap |
|---|---|---|---|
| Calcium derivative | Native | Calcium derivative | |
| Data collection statistics | |||
| Diffraction Sources | Synchrotron CLS08ID-1 | Synchrotron CLS08ID-1 | Home |
| Wavelength (Å) | 1.77123 | 0.97949 | 2.29097 |
| No. of molecules in asymmetric unit | one | two | two |
| Space group | C121 | P1 | P1 |
| a, b, c (Å) | 202.8, 45.6, 58.6 | 47.7, 50.4, 124.4 | 48.0, 50.4, 123.2 |
| α, β, γ (°) | 90.0, 100.0, 90.0 | 97.6, 98.2, 101.8 | 96.9, 98.2, 102.1 |
| Resolution range (Å)* | 46.6 – 2.0 (2.05 – 2.0) | 48.64 – 1.75 (1.78–1.75) | 48.65 – 2.5 (2.61 – 2.51) |
| Rmerge* | 0.10 (0.47) | 0.08 (0.83) | 0.07 (0.17) |
| Rmeas * | 0.11 (0.49) | 0.09 (0.96) | 0.08 (0.20) |
| CC 1/2* | 0.999 (0.990) | 0.998 (0.859) | 0.997 (0.985) |
| < I/σ > (I) >* | 19.6 (4.2) | 10 (1.9) | 16.9 (6.7) |
| Completeness (%)* | 99.0 (87.6) | 97.3 (95.3) | 91.9 (87.9) |
| Redundancy* | 14.1 (11.4) | 3.9 (4.0) | 3.9 (3.9) |
| Refinement statistics | |||
| Resolution range (Å) | 46.6 – 2.0 | 48.64 – 1.75 | |
| No. of reflections | 34,009 | 103,273 | |
| Rwork/Rfree | 018/0.23 | 0.18/0.21 | |
| No. of atoms | |||
| Protein | 2638 | 6050 | |
| CA ion | 4 | 26 | |
| Water | 498 | 974 | |
| Overall B-factors from Wilson plot (Å2) | 28.2 | 17.9 | |
| Average B-factor (Å2) | |||
| Protein | 29.6 | 13.7 | |
| Water | 43.6 | 35.3 | |
| CA ion | 27.9 | 24.4 | |
| R.m.s. deviations | |||
| Bond length (Å) | 0.011 | 0.011 | |
| Bond angle (°) | 1.726 | 1.574 | |
| Ramachandran plot statistics (%) | |||
| Most favoured region | 96.6 | 97.28 | |
| Additionally allowed regions | 3.11 | 2.72 | |
| PDB ID | 6XI3 | 6XI1 | |
* The numbers in parentheses are for the highest resolution shell.
2.4. Molecular dynamics
Molecular dynamics simulations of MhLap and AhLap di-tandemers were undertaken as previously described (Hakim et al., 2013). Briefly, GROMACS was used to place each di-tandemer in a box of virtual waters and perform energy minimization. This was followed by constant-volume and constant-pressure position-restrained molecular dynamics runs, each 0.1 ns long. Unrestrained molecular dynamics runs were then performed, lasting 20 ns. The duration of this simulation is likely not long enough to see the unfolding of protein domains but is long enough to observe relative changes in domain orientation (Mayor et al., 2000). All simulations were run with a fixed temperature of 298 K. The same protocol was followed for di-tandemers where the calcium ions had been removed.
The output topology and trajectory files were loaded into VMD to analyze the positions of the beta sandwiches relative to each other, and how this changed over time. To measure potential bending of the linker between the domains (Peters et al., 2017), an oblique triangle was drawn between three atoms (MhLap = Leu240Cβ – Val247Cβ – Ile158Cβ, AhLap = Ile228Cβ – Val264Cβ – Tyr300Cβ), and the obtuse angle was measured at each frame. To measure potential twisting of the linker between the domains, three lines were drawn between four atoms (MhLap = Val294Cα – Leu240Cβ – Ile158Cβ – Ser130Cα, AhLap = Ile334Cα – Tyr300Cβ – Ile228Cβ – Glu152Cα), and the dihedral angle of the system was measured at each frame.
3. Results
3.1. Sequence analysis of RTX adhesin extender regions
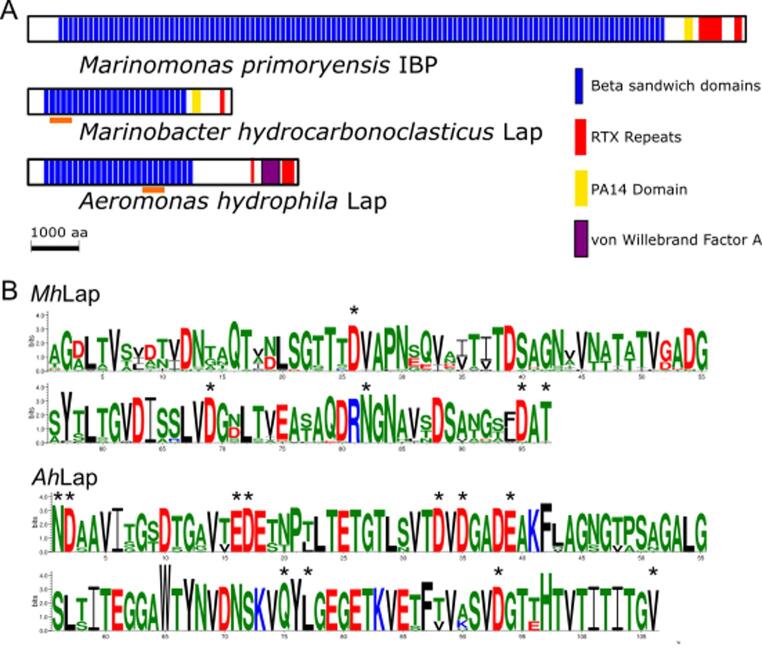
One of the obvious differences in the architectures of the full-length adhesins AhLap, MhLap, and the previously characterized MpIBP, is in the number of repeats present in the extender region (Fig. 1A). While the MpIBP has a predicted 120 repeats (which makes up 90% of its 1.5-MDa molecular weight) (Guo et al., 2012), the MhLap and AhLap are much shorter, with only ~ 20 repeats each. RTX adhesins with repeat numbers between 20 and 120 are plentiful in the NCBI database, including examples such as LapA (~40), FrhA (~10), SiiE (~50), and RtxA (5 – 40, depending on the strain analysed). It should be noted that adhesin repeat numbers are often underrepresented in genome databases. This occurs when sequencing reads are unable to stretch across large areas of highly repetitive sequence, leading to genome assemblies that fail to show the proper number of repeats. Several such cases have been reported (D’Auria et al., 2008, Guo et al., 2017, Wrobel et al., 2018), and likely many more have yet to be caught. For instance, only a single repeat is reported in the AhLap sequence from the NJ-35 strain (Dong et al., 2020), which could be an underestimate.
Fig. 1.
RTX adhesin domain architecture. (A) The domain architectures for three exemplar RTX adhesins are shown, with their N termini on the left. Beta sandwiches and RTX repeats – domains that appear in all known RTX adhesins – are coloured blue and red, respectively. Two known domains used for bacterial adhesion are PA14 and vWFA, coloured yellow and purple, respectively. The repeats used for the tetra-tandemer constructs in this study are underlined in orange. (B) Weblogos for all the beta-sandwich repeats present in MhLap or AhLap. The residues are colour-coded red for negatively charged, blue for positively charged, black for hydrophobic, and green for polar uncharged/small aliphatic. Asterisks are above calcium-binding residues.
Zeroing in on the sequence of each repeat, the lengths of the beta sandwiches average around 100 amino acids, but the sequence identity between repeats varies wildly. Beta sandwiches within MpIBP share 100% sequence identity even at the DNA level, but the MhLap and AhLap repeats show greater variability, with repeat identity ranging between 65 and 90 % and 85 to 95% at the amino-acid level, respectively. A web logo plot of the MhLap and AhLap beta sandwiches shows this variability, with MhLap repeats clearly varying more than those of AhLap (Fig. 1B). In both cases, a high number of negatively charged aspartate and glutamate residues, as well as their amide derivates, asparagine and glutamine, are present and highly conserved throughout the repeats. Few positively charged residues are present in either protein, although the lysines and arginines that are present are highly conserved.
Taking these sequence observations into account, tetra-tandemer constructs were chosen to maximize the presence of conserved residues, while also providing a spread in sequence identity comparable to the adhesin as a whole (Table 2). Fig. 1A shows from where the tetra-tandemer sequences were drawn within the full-length adhesins, namely towards the N-terminal end for MhLap and the in latter third of the repeats for AhLap. Genes encoding these segments were synthesized, using codon redundancy to keep the amino-acid sequence the same as the native protein (Fig. S1), while reducing the sequence identity of the repeats at the DNA level to below 75%, thereby reducing the chance of DNA recombination in E. coli (Bzymek and Lovett, 2001).
Table 2.
Sequence identity of the beta-sandwich repeats within and between RTX adhesins.
| MpIBP | MhLap | AhLap | |
|---|---|---|---|
| MpIBP | 100% | 28–36% | 19–22% |
| MhLap | 28–36% | 77–87% | 15–23% |
| AhLap | 19–22% | 15–23% | 94–98% |
3.2. Tetra-tandemers show calcium-related structure changes of varying magnitudes
Both the MhLap and AhLap tetra-tandemers were expressed in E. coli and purified through a combination of nickel-affinity and size-exclusion chromatography. From lysis onwards, both proteins were kept in 2 mM CaCl2, in case these tetra-tandemers – like MpIBP – require calcium for proper folding. To test this possibility, the pure proteins were dialyzed against EDTA to remove ambient and bound calcium, and then dialyzed back into a calcium-deprived version of its initial buffer. A calcium titration was then undertaken, with secondary structure changes being monitored through circular dichroism (CD) spectroscopy (Fig. 2).
Fig. 2.
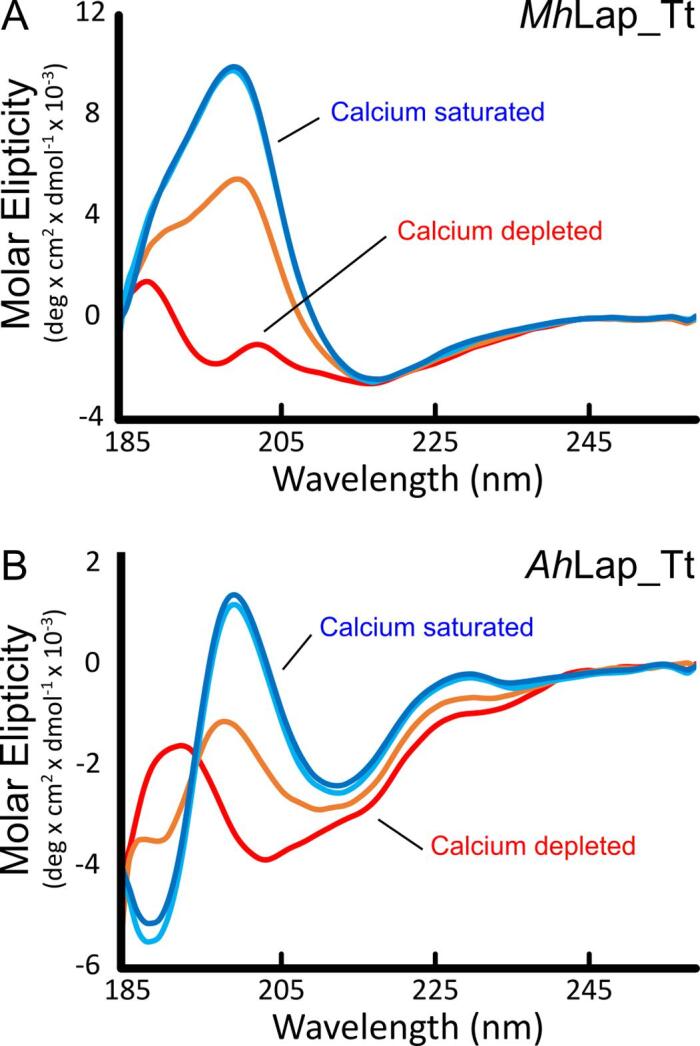
Circular dichroism spectroscopy. The CD spectra of a calcium titration for the MhLap (A) and the AhLap (B) tetra-tandemers. The titrations began in 0.01 mM EDTA (red), with subsequent additions of CaCl2 injected to produce ambient concentrations of 0.5 mM Ca2+ (orange), 1 mM Ca2+ (cyan) and 2 mM Ca2+ (dark blue).
In the absence of calcium, the MhLap tetra-tandemer spectrum (red line) shows relatively small changes in molar ellipticity from the baseline, with two minor peaks at 189 and 202 nm, and a minimum at 218 nm (Fig. 2A). Such a spectrum is not easily interpreted as being dominated by a particular secondary structure, instead it suggests there is a roughly even mixture of random coil and beta strand. Addition of 0.5 mM CaCl2 led to a drastic change in the spectrum, with a single large maximum appearing at 200 nm (orange line), while its initial minimum at 215 nm is maintained. This change was enhanced by doubling the CaCl2 concentration to 1 mM (cyan line), but did not change with further additions of CaCl2, indicating a saturation in Ca2+-binding. Taken together, these spectra demonstrate that the MhLap tetra-tandemer develops into a beta-strand-dominated structure upon Ca2+ addition, although some structure is present in the absence of calcium.
The AhLap tetra-tandemer revealed a similar transition from less to more beta structure upon calcium addition, once again saturating at 1 mM CaCl2 (Fig. 2B). However, the shape of the AhLap spectra, as compared to those of MhLap, was rather different, with the spectrum of the fully-folded AhLap tetra-tandemer (cyan and dark blue lines) featuring two peaks (major at 200 nm, minor at 230 nm) and two troughs (major at 189 nm, minor at 213 nm), all at much lower molar ellipticity values. Deconvolution of the spectra indicated a lower percentage of beta strand for AhLap (37%) than for MhLap (49%) (Table 3), suggesting that the two take on somewhat different folds once calcium-bound, at least at the secondary structure level.
Table 3.
Deconvolution from CD spectra of MhLap and AhLap tetra-tandemers.
|
MhLap |
AhLap |
|||||
|---|---|---|---|---|---|---|
| 0 mM Ca2+ | 1 mM Ca2+ | Structure | 0 mM Ca2+ | 1 mM Ca2+ | Structure | |
| Alpha helix | 3% | 3% | 0% | 0% | 0% | 0% |
| 310 helix | 0% | 0% | 0% | 4% | 2% | 5% |
| Beta strand | 37% | 49% | 66% | 34% | 37% | 55% |
| Turns | 18% | 22% | 26% | 17% | 21% | 28% |
| Unordered | 41% | 26% | 8% | 45% | 39% | 12% |
3.3. Tetra-tandemers share similar macro-structure, while the specifics of the folds differ
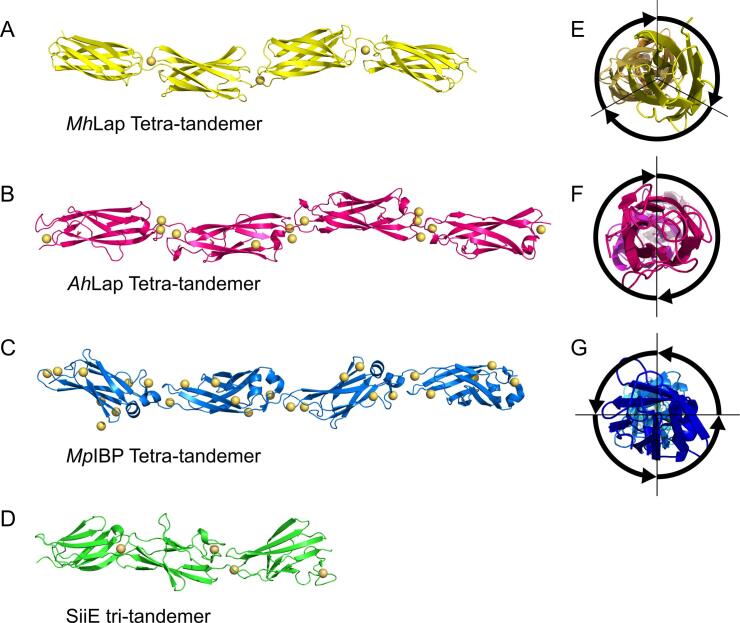
To interrogate the apparent structural difference between the tetra-tandemers, both were crystallized under similar conditions, yielding crystals with asymmetrically elongated unit cells (Table 1). Calcium SAD phasing (Guo et al., 2019a, Guo et al., 2019b) was used to solve the tetra-tandemer structures to 2.0 Å (MhLap) and 1.75 Å resolution (AhLap). These structures confirm that the MhLap (Fig. 3A) and AhLap (Fig. 3B) tetra-tandemers are made up of four tandem beta-sandwich-like domains that are strung together by short linkers into a linear, rod-like arrangement within the crystal, reminiscent of the previously-solved MpIBP tetra-tandemer (Fig. 3C) and, to a lesser extent, the SiiE tri-tandemer (Fig. 3D). The repeats are rotated relative to each other around the long central axis, producing a handedness to the tetra-tandemers reminiscent of helices. Both the MhLap (Fig. 3E) and AhLap (Fig. 3F) structures show right-handed rotations, with pitches of approximately i + 3 and i + 2, respectively. As previously shown, the MpIBP tetra-tandemer rotation stands in contrast as left-handed (Fig. 3G) and can complete a full rotation in five repeats (i + 4). The handedness of SiiE is harder to define, as there are only three domains, the first being distinct in shape and sequence from the others (Griessl et al., 2013).
Fig. 3.
Tetra-tandemer structures. Crystal structures of the (A) MhLap, PDB code: 6XI3 (this study), (B) AhLap, PDB code: 6XI1 (this study), and (C) MpIBP, PDB code: 4P99 (Vance et al., 2014) tetra-tandemers, as well as the SiiE tri-tandemer, PDB code: 2YN3 (Griessl et al., 2013). Calcium ions are shown as gold spheres. The repeats rotate down a central axis of varying periodicity and direction. Views down that axis from N to C terminus are shown for MhLap (D), AhLap (E), and MpIBP (F), along with their direction and the relative position of repeats.
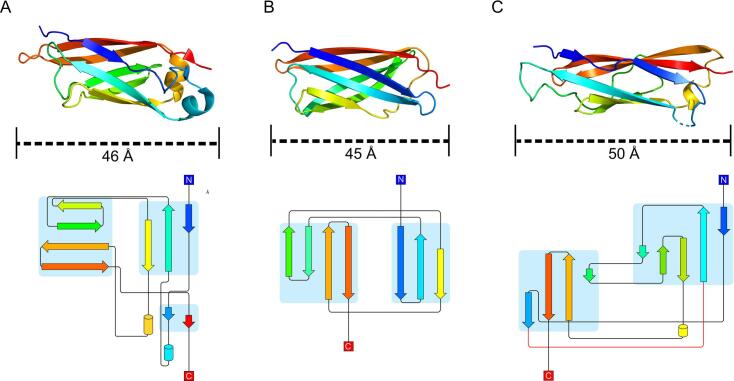
Looking closer at the individual repeats for each tetra-tandemer, more differences between the beta sandwiches become apparent. Using the MpIBP as a reference, the 46-Å long repeats are mainly comprised of two antiparallel beta sheets – one three-stranded, the other four-stranded – along with a short, two-stranded, parallel sheet and two alpha helices at the C-terminal end (red) of the domain (Fig. 4A). The 45 Å-long MhLap repeats are similar yet simpler (Fig. 4B), keeping the two antiparallel beta sheets but lacking the ancillary structural elements present in MpIBP (i.e. the short sheet, and the helices). Despite these differences, the connectivity and partnering of the strands is conserved. Alternatively, the AhLap repeats possess a more involved connectivity, resulting in 50 Å-long beta sandwiches that are held together by one four-stranded and another five-stranded beta sheet (Fig. 4C). Interestingly, the second strand and the final strand hydrogen bond in a parallel fashion, similar to the short sheet at the C terminus of the MpIBP repeats, while the rest of the strands are antiparallel. As another point of comparison, the beta sandwiches from all three tetra-tandemers are longer than the three solved SiiE beta sandwiches, which range from 30 to 35 Å (Fig. S2). The SiiE extender region is known to possess two distinct beta sandwiches, typified here by Ig50 and Ig51. The two beta sandwiches show remarkably different connectivities, with Ig50 being more similar to the simpler MhLap (Fig. S2).
Fig. 4.
Structural comparison of beta-sandwich monomers. Crystal structure and domain topology diagrams for MpIBP (A), MhLap (B), and AhLap (C). Structures are coloured by primary sequence to progress from N terminus (blue) to C terminus (red); the strands (arrows) and helices (cylinders) in the topology diagrams (below) are similarly coloured, with the connecting loops in black (or red if not observed in the crystal structure). The length of each monomer is denoted by the black dashed lines below the structures.
Even with the differences in domain folding and relative rotation, the tetra-tandemers (and SiiE) all maintain a similarly elongated conformation. This conservation of macro-structure is in spite of the seemingly flexible interstitial loops between domains, which lack defined secondary structure. All four structures reveal coordinated Ca2+ within these regions (Fig. 3), varying in number and coordination pattern, yet apparently conserved in purpose: to rigidify the linkers and project an elongated conformation.
3.4. MhLap coordinates a single Ca2+ within the flexible linker regions
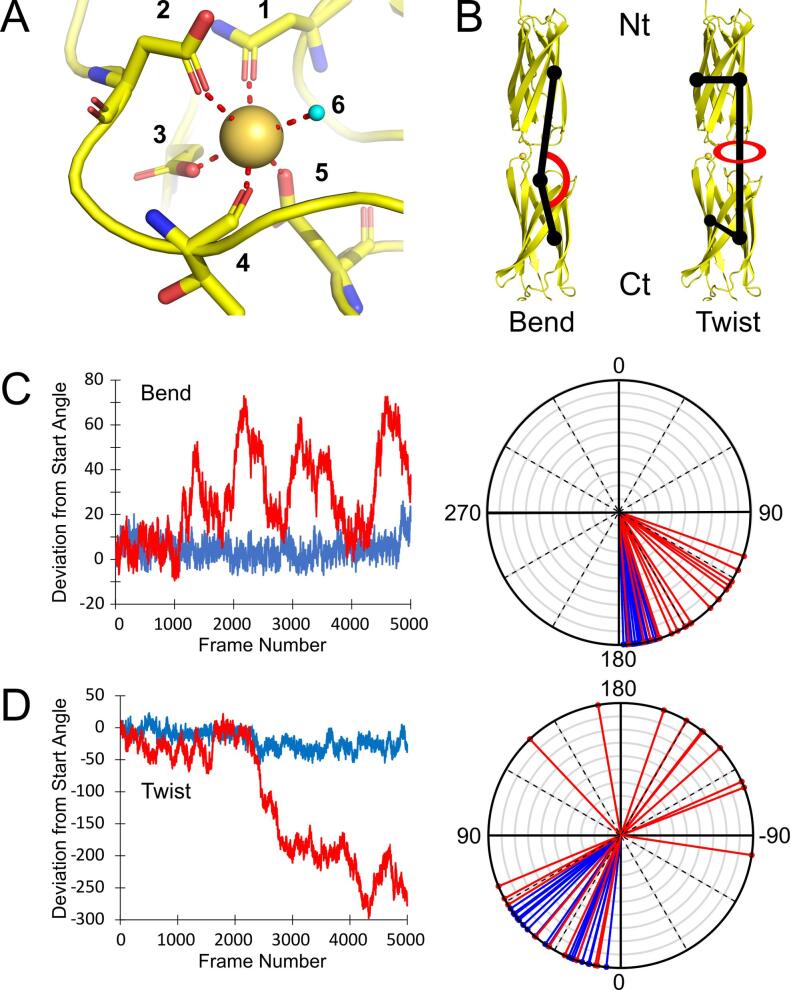
Regarding Ca2+ coordination, the MhLap tetra-tandemer is once again the simplest of the three. While the MpIBP tetra-tandemer was found to have many Ca2+ coordinated throughout its structure, MhLap consigns its calcium ions to the linker regions, with only one ion per domain interface. This limited calcium complement may explain the differences seen in CD spectra during calcium titrations, as MpIBP tetra-tandemer loses all beta and alpha characteristics in EDTA (Table S1) (Guo et al., 2013), while MhLap retains some level of structure (Fig. 2A).
Each calcium in the MhLap tetra-tandemer structure is bound in a similar manner (Fig. 5), forming an octahedral coordination sphere comprised of five protein contacts and one water molecule. The protein contacts are made by the sidechains of one asparagine and three aspartate residues, as well as a single backbone carbonyl (Fig. 5A). These Asn/Asp residues are conserved throughout the entirety of the full-length MhLap extender region (Fig. 1B), though there are many other like residues that are equally-well conserved that do not appear to be involved in calcium coordination.
Fig. 5.
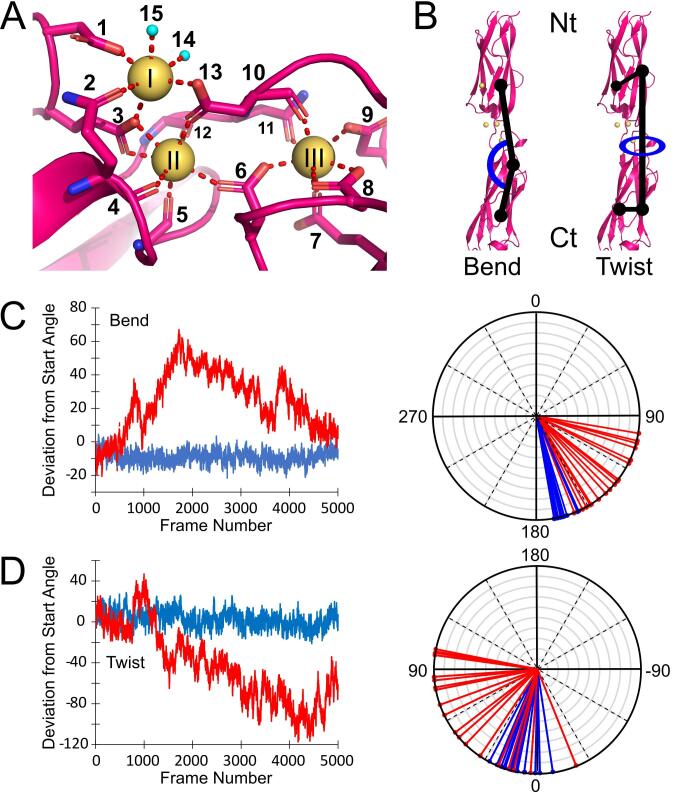
MhLap calcium coordination and molecular dynamics. (A) The coordination of inter-repeat Ca2+ in the MhLap tetra-tandemer. Ca2+ is shown as a gold sphere, and residues binding to it are numbered as follows: 1 = Asn205, 2 = Asp121, 3 = Asp95, 4 = Thr123, 5 = Asp149, 6 = water. Water molecules participating in the coordination sphere are shown as cyan spheres. (B) The di-tandemer in silico construct used for molecular dynamics simulations. The Bend and Twist angles to be monitored throughout the simulations are shown, and the termini are labelled Nt and Ct. (C) The Bend angle throughout the simulations, depicted in two ways. Left: a plot of deviation from start angle over frame number. Right: a radial plot where the Bend angle every 250 frames is denoted by a line. Simulations with (blue) and without (red) the coordinated Ca2+ ions are shown. (D) The Twist angle throughout the simulations, depicted the same way as the Bend angle.
To analyze the potential rigidifying properties of these interstitial calcium ions, molecular dynamics simulations on a di-tandemer of the MhLap were undertaken with and without the Ca2+ present. Two angles were measured throughout the 5000 frames (Fig. 5B), one to assay bending of the domains at the linker region (Bend), the other to assay rotation of the repeats about the central axis (Twist). In the presence of calcium, both angles underwent minor changes over the course of the simulation (blue tracing in Fig. 5C and D). The di-tandemer maintained a rod-like structure, with the Bend angle between the two domains never deviating over 30° relative to the start frame; the two domains saw slightly more twisting over the simulation, maintaining a ~ 60° wedge.
Removing the calcium introduced major variability in both angles. With regards to bending, the two domains oscillated between an angle close to that in the start frame and a second angle ~ 70° from the start frame (red tracing in Fig. 5C, left), bending back and forth over the course of the 20 ns simulation four times. At points, the two domains approach a perpendicular orientation relative to each other (Fig. 5C, right). The twist dihedral angle drastically changes from the start frame to approach a ~ 300° deviation (Fig. 5D, left). Indeed, the two domains can sample almost the entire 360° rotation throughout the simulation (Fig. 5D, right). Visualizing the protein at select frames makes the magnitude of these structural changes strikingly clear (Fig. 6). The di-tandemer at frame 1000 (blue) is very similar to the crystal structure’s orientation (light gray). But as the simulation progresses, the two beta sandwiches sample a wide variety of relative angles (cyan through red). The aforementioned perpendicular orientation can be seen at frame 2200 (cyan).
Fig. 6.
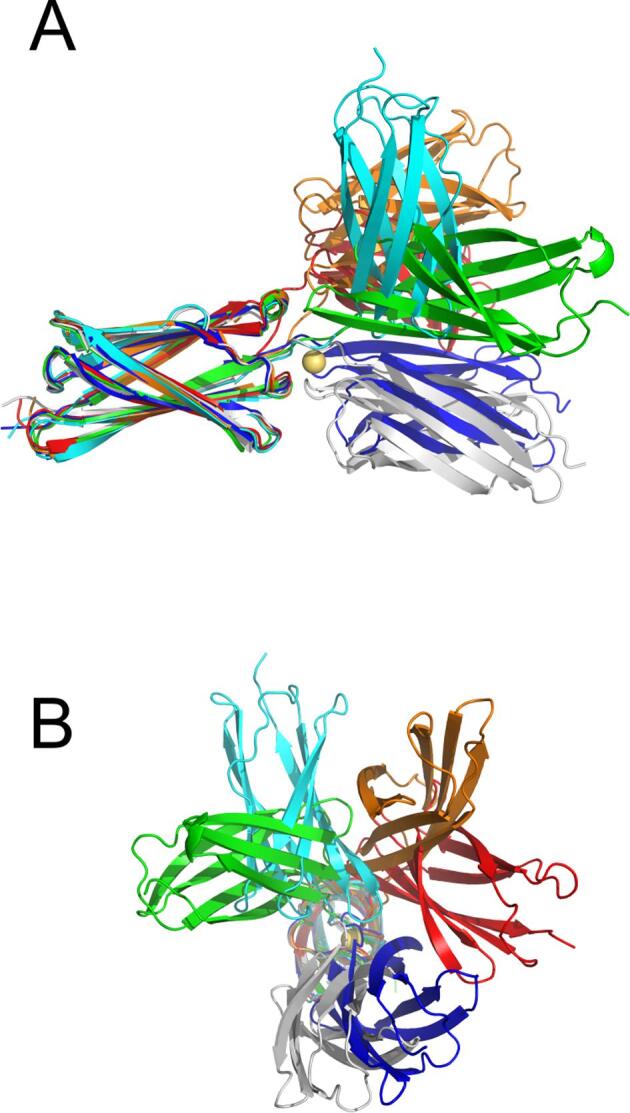
Frames from the MhLap No-Ca2+ molecular dynamics simulation. The first repeat of the di-tandemer construct at frames 1000 (blue), 2200 (cyan), 3000 (green), 4000 (orange), and 4500 (red) were aligned against the first repeat of the truncated crystal structure (white). Both a side view (A) and a view down the central axis (B) are shown.
3.5. AhLap uses three Ca2+ to maintain its rod-like conformation
The AhLap tetra-tandemer coordinates many more calcium ions throughout its length than does MhLap, although the number of ions per repeat is inconsistent. Each inter-domain interface contains, at minimum, three highly coordinated Ca2+ (Fig. 7A), labelled as calciums I, II, and III. Of the three, calcium I has the fewest coordinate bonds to protein-based ligands, with two positions in its six-ligand octahedral coordination sphere being taken up by water molecules. Both calciums II and III have pentagonal bipyramid coordination spheres, comprised of seven protein-based ligands each. The three Ca2+ are all coordinated by a combination of Asp, Asn, and Glu sidechains that are conserved in every repeat of the full-length adhesin (except the Glu in position 7, which is a comparable Gln for five repeats), as well as backbone carbonyl groups (Table 4). Interestingly, several acidic residues (namely ligands 3, 6, and 13 in Fig. 7A) use their bidentate sidechains to coordinate two separate calcium ions at the same time, making calciums I, II and III part of an interconnected coordination network.
Fig. 7.
AhLap calcium coordination and molecular dynamics. (A) The coordination of inter-repeat Ca2+ in the AhLap tetra-tandemer. Ca2+ are shown as gold spheres, and residues binding to them are numbered as follows: 1 = Asp144, 2 = Gln202, 3 = Glu143, 4 = Gln202, 5 = Leu204, 6 = Asp268, 7 = Glu272, 8 = Asp266, 9 = Asp326, 10 = Asp235, 11 = Asn234, 12 = Val233, 13 = Asp235, 14 = water, 15 = water. Water molecules participating in the coordination sphere are shown as cyan spheres. (B) The di-tandemer in silico construct used for molecular dynamics simulations. The Bend and Twist angles to be monitored throughout the simulations are shown, and the termini are labelled as in Fig. 5. (C) The Bend angle throughout the simulations, depicted in two ways. Left: a plot of deviation from start angle over frame number. Right: a radial plot where the Bend angle every 250 frames is denoted by a line. Simulations with (blue) and without (red) the coordinated Ca2+ ions are shown. (D) The Twist angle throughout the simulations, depicted the same way as the Bend angle.
Table 4.
Coordinating ligands of inter-domain Ca2+ ions.
|
MhLap Tetra-tandemer |
AhLap Tetra-tandemer |
||||
|---|---|---|---|---|---|
| Residue | Number | Residue | Number | ||
| 1 | Asn sidechain | 205/302/399 | 1 | Asp sidechain | 38/144/250 |
| 2 | Asp sidechain | 121/218/315 | 2 | Gln sidechain | 96/202/308 |
| 3 | Asp sidechain | 95/192/289 | 3 | Glu sidechain | 37/143/249 |
| 4 | Thr backbone | 123/220/317 | 4 | Gln backbone | 96/202/308 |
| 5 | Asp sidechain | 149/246/343 | 5 | Leu backbone | 98/204/310 |
| 6 | Water | 6 | Asp sidechain | 162/268/374 | |
| 7 | Glu sidechain | 166/272/378 | |||
| 8 | Asp sidechain | 160/266/372 | |||
| 9 | Asp sidechain | 220/326/432 | |||
| 10 | Asp backbone | 129/235/341 | |||
| 11 | Asn sidechain | 128/234/340 | |||
| 12 | Valbackbone | 127/233/339 | |||
| 13 | Asp sidechain | 129/235/341 | |||
| 14 | Water | ||||
| 15 | Water | ||||
Besides these three consistent calcium ions, additional Ca2+ are present in select repeats. The AhLap structure was solved with two tetra-tandemers in the asymmetric unit, meaning that the structure holds eight repeat domains with six replicate interfaces between them. Of those six, four of them hold an additional calcium (Fig. S3). These calcium ions are held in place by only two protein-based ligands, with the rest of the octahedral coordination sphere made up of water ligands. This minimal connection to the protein explains the inconsistent presence of the ion in the structure. A single additional calcium is found outside of the domain interfaces, attached to the side of the second repeat in chain A. As expected, this calcium has fewer protein contacts, and is only coordinated by two protein-based ligands (Fig. S4). Whether these additional ions are present in vivo is uncertain, though the residues that coordinate them are highly conserved throughout the entirety of AhLap.
Molecular dynamics simulations were run on an AhLap di-tandemer in the presence and absence of calcium, as was done for MhLap. Again, two similar angles were measured throughout to observe the relative twisting and bending of the domains (Fig. 7B). Only the three conserved calcium ions were included in the calcium-containing run, which produced an expected minimal amount of bending or twisting between the domains (blue tracing in Fig. 7C and D). In fact, the AhLap tetra-tandemer appears more rigid than the MhLap, with both angles remaining closer to the start angle over the entirety of the run.
Removal of the conserved calcium ions once again led to a stark increase in variability for both angles. The two domains were able to bend at the linker region to reach a 70° deviation from the start angle (red tracing in Fig. 7C, left) almost becoming perpendicular to each other at times (Fig. 7C, right). Interestingly, this change was less frequent than for MhLap, with a single oscillation taking the whole simulation, while MhLap underwent several throughout its run. The twisting between the AhLap domains was not as severe as for MhLap, deviating by a striking 120° (Fig. 5D), which is far less than MhLap’s ~ 300° wedge. Aligning select frames once again showcases the variety of orientations taken up by the di-tandemer (Fig. 8), especially in frames 1000 (cyan) and 4000 (orange) where the repeats are almost perpendicular to each other. This comparison also visualizes how the AhLap di-tandemer is apparently less flexible than MhLap, limiting the orientations it can explore.
Fig. 8.
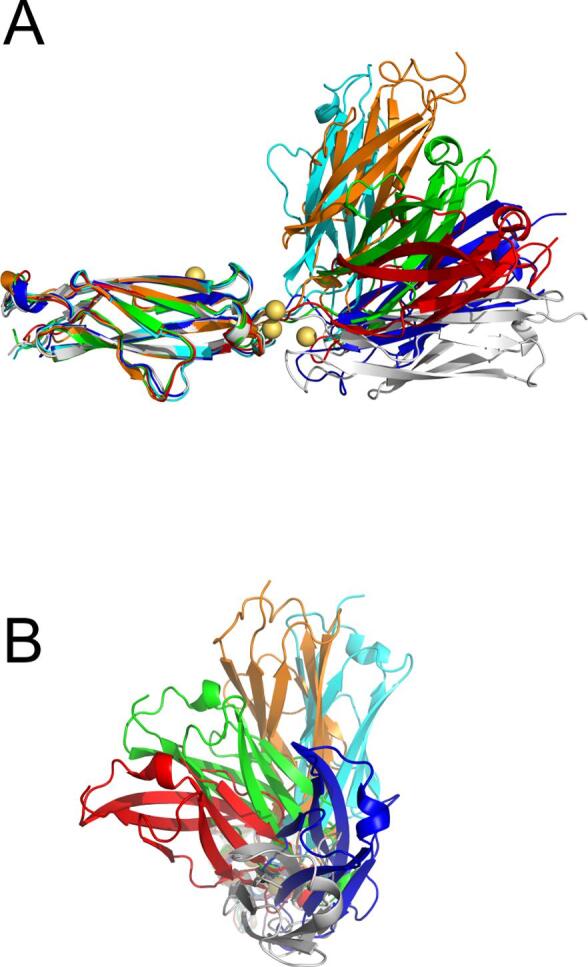
Frames from the AhLap No-Ca2+ molecular dynamics simulation. The first repeat of the di-tandemer construct at frames 500 (blue), 2000 (cyan), 3000 (green), 4000 (orange), and 5000 (red) were aligned against the first repeat of the truncated crystal structure (white). Both a side view (A) and a view down the central axis (B) are shown.
In summary, a similar trend is observed from both simulations: the absence of calcium removes the stable rod-like structure of these tandem beta-sandwich repeats, leading to a much greater freedom in domain orientation.
4. Discussion:
4.1. Calcium coordination as a prerequisite for RTX-mediated biofilms
Recently, Dong et al. published a paper detailing hyper biofilm-forming mutants in A. hydrophila strains that infect fish (Dong et al., 2020). It was found that an overproduction of AhLap (named against convention within the study as RmpA) was responsible for this new phenotype, adding AhLap to the growing list of RTX adhesins vital to biofilm formation (Guo et al., 2017, Hinsa et al., 2003, Martínez-Gil et al., 2010, Syed et al., 2009). While deletions of AhLap removed the hyper biofilm-forming phenotype as expected, so too did the removal of Ca2+ from the solution via chelating agents (Dong et al., 2020). This result mirrors other such experiments on RTX adhesin-mediated behaviour, like studies on the RTX adhesin SiiE in Salmonella enterica, where the removal of calcium greatly reduced invasion of the bacteria into polarized epithelial cells (Peters et al., 2017).
The experiments on the auto-aggregating AhLap mutants were unable to pinpoint exactly what effect Ca2+ removal had on the adhesin-mediated biofilm. Fortunately, previous research into the biochemistry of RTX proteins (including this study) can offer some suggestions. Studies have shown that the relationship between RTX adhesins and Ca2+ is a complex and multifaceted one, impacting proper secretion, folding, orientation, and adhesin-substrate contacts. As indicated by CD spectroscopy results both here and elsewhere (Guo et al., 2017, 2013), coordinated calcium ions are key in the folding of most RTX adhesin domains, which impacts everything from the protease-resistance of the adhesins (Peters et al., 2017) to their localization on the cell surface via the type 1 secretion system (T1SS). T1SS-secreted proteins must remain unfolded for transport through the dual membranes of Gram-negative bacteria, and while there are some that require chaperone proteins (Delepelaire and Wandersman, 1998), it is thought that the adhesins rely on calcium-dependent folding to remain intracellularly unfolded (Bumba et al., 2016). Interestingly, our results show that certain domains from RTX adhesins are not completely unfolded in the absence of Ca2+, as have previous studies on domains from MhLap (Vance et al., 2019) and SiiE (Peters et al., 2017). The calcium-free partial structures of such RTX adhesin domains are likely unstable and weakly held together, as was shown in studies on the thermal stability of calcium-free mutants of SiiE beta sandwiches (Peters et al., 2017) and single-molecule force microscopy experiments on calcium-deprived MpIBP octa-tandemers (Oude Vrielink et al., 2017). As such, these intracellular structures are expected to easily pull apart during secretion through the pores of the T1SS, refolding into their strong calcium-bound structures as they reach the extracellular space.
Examination of the adhesion domains from MhLap (Vance et al., 2019) and MpIBP (Guo et al., 2017) have demonstrated how Ca2+ is also at the forefront of adhesin-substrate interactions, where the ions coordinate ligands from both adhesin and glycan/peptide substrates. However, these contacts could not be established if calcium ions were not also involved in determining the proper orientation of the adhesin’s domains relative to each other in 3-D space. While inter-domain calcium ions are likely present throughout the adhesin, perhaps orienting the adhesion domains in the C-terminal regions into particular macrostructures (Guo et al., 2017, Guo et al., 2019a, Guo et al., 2019b), certainly this Ca2+-mediated ordering of domains is most obvious in the rigidification of the extender region. Demonstrated clearly by the MD simulations of AhLap in the absence of calcium, the adhesin’s stalk could not channel the exploration of the adhesion domains to substrates away from the bacterium’s own surface if the domains could bend and twist as freely as they do in the absence of Ca2+.
4.2. Calcium-induced rigidification: A conserved or convergent strategy?
The importance of a rigid extender region is highlighted by their presence throughout adhesin families, both RTX and otherwise. Filamentous adhesins, such as the type I pili from E. coli, connect many Ig-like monomers into long threads that extend the sugar-binding tip towards a substrate (Schilling et al., 2001). The type Ve autotransporter adhesins from Gram-negative bacteria also use beta-sandwich extenders (Tsai et al., 2010), as do the Gram-positive biofilm-associated proteins (Baps) (Cucarella et al., 2001). Yet, while rigidification appears to be a prerequisite for all useful extension regions, the use of calcium-induced rigidification is not universal. For example, neither pili nor autotransporter adhesins require calcium for rigidification. Instead, type I pili assemble their monomers into a stiff helical shape (Hospenthal et al., 2017, Sauer et al., 2004), and invasins use linkers rich in proline residues (Palumbo and Wang, 2006).
Is calcium rigidification conserved throughout all RTX adhesins? Our studies here have increased the number of linked extender region structures in RTX adhesins from two to four. In all four structures, the beta sandwiches coordinate calcium ions in the linker regions between domains. The weight of evidence suggests that these calcium ions are used for rigidification, as seen for SiiE by electron microscopy, small angle x-ray scattering (SAXS), and molecular dynamics simulations (Griessl et al., 2013, Peters et al., 2017); for MpIBP by SAXS and single-molecule force microscopy (Oude Vrielink et al., 2017, Vance et al., 2014), and now for both the MhLap and the AhLap by molecular dynamics. Beyond the studied examples, sequences of predicted RTX adhesins within the NCBI database show a consistent excess of negatively charged residues, suggesting that this calcium-rigidification strategy is widely conserved.
However, while the outcome remains the same, the specifics of how calcium stabilizes the rod-like structure of these tandem repeats vary. Between the three tetra-tandemers and the heavily studied SiiE tri-tandemer, both the number of calcium ions present between repeats and the coordinating residues of these ions are remarkably different. Along with the low sequence identity between different adhesin’s beta-sandwich domains, one begins to question whether this calcium-rigidification strategy is really conserved from a progenitor adhesin in all cases, or – as an alternative theory –the strategy arose convergently between different species, finding similar solutions to the need for rigidification and extension but via different origins. The profound differences in the AhLap monomer (both sequence identity and connectivity) and calcium coordination relative to both MpIBP and MhLap would seem to support evolutionary convergence.
5. Conclusions
Expansive extender regions made up of tandem beta-sandwich repeats are a defining feature of the biofilm-associated RTX adhesins. Here, segments of extender regions – one from an oil-eating bacterium, the other an opportunistic pathogen of fish and mammals – were structurally characterized. In keeping with the known importance of calcium for RTX adhesins, both extender segments were found to coordinate Ca2+ in a manner that maintains the otherwise unwieldy string of beta sandwiches as a rigid and rod-like structure. However, there were striking differences in the structure and coordination strategy between these proteins, raising questions as to the evolutionary origin of these regions in supposedly homologous adhesins across the bacterial domain.
Research into this expanding family of adhesins continues to show the importance of calcium, and the specifics of how the ions are integrated into the extracellular protein’s functions are becoming more complicated. As strategies to control RTX adhesin-mediated biofilms mature, the keystone calcium ion will become an obvious target. While no one solution for all biofilms is likely to exist, the dependence of RTX adhesins on calcium as a potent connector may come close.
6. Data availability
X-ray crystal structure coordinates solved in this study have been deposited in the Protein Data Bank with accession codes 6XI3 (MhLap) and 6XI1 (AhLap). Data that support the findings of this study are available from the corresponding author P.L.D. upon reasonable request.
CRediT authorship contribution statement
Tyler D.R. Vance: Conceptualization, Software, Investigation, Visualization, Supervision. Qilu Ye: Validation, Investigation. Brigid Conroy: Investigation. Peter L. Davies: Conceptualization, Resources, Supervision, Project administration, Funding acquisition.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
The authors wish to acknowledge Sherry Gauthier for her assistance with molecular cloning, and Kim Munro from Queen’s University’s Protein Function Discovery facility for his assistance with circular dichroism spectroscopy. We are grateful to Dr. John Allingham for access to his home X-ray source diffractometer. Structural data were also collected with help from staff at the Canadian Light Source (Saskatoon, SK); and molecular dynamics simulations were run using CPU clusters of the High-Performance Computing Virtual Laboratory (Kingston, ON).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.yjsbx.2020.100036.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- Afonine P.V., Grosse-Kunstleve R.W., Echols N., Headd J.J., Moriarty N.W., Mustyakimov M., Terwilliger T.C., Urzhumtsev A., Zwart P.H., Adams P.D. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr D. 2012;68:352–367. doi: 10.1107/S0907444912001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boggon T.J., Murray J., Chappuis-Flament S., Wong E., Gumbiner B.M., Shapiro L. C-Cadherin Ectodomain Structure and Implications for Cell Adhesion Mechanisms. Science. 2002;296:1308–1313. doi: 10.1126/science.1071559. [DOI] [PubMed] [Google Scholar]
- Bork P., Holm L., Sander C. The Immunoglobulin Fold: Structural Classification, Sequence Patterns and Common Core. J. Mol. Bio. 1994;242:309–320. doi: 10.1006/jmbi.1994.1582. [DOI] [PubMed] [Google Scholar]
- Borlee B.R., Goldman A.D., Murakami K., Samudrala R., Wozniak D.J., Parsek M.R. Pseudomonas aeruginosa uses a cyclic-di-GMP-regulated adhesin to reinforce the biofilm extracellular matrix. Mol. Microbiol. 2010;75:827–842. doi: 10.1111/j.1365-2958.2009.06991.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bumba L., Masin J., Macek P., Wald T., Motlova L., Bibova I., Klimova N., Bednarova L., Veverka V., Kachala M., Svergun D.I., Barinka C., Sebo P. Calcium-Driven Folding of RTX Domain β-Rolls Ratchets Translocation of RTX Proteins through Type I Secretion Ducts. Mol. Cell. 2016;62:47–62. doi: 10.1016/j.molcel.2016.03.018. [DOI] [PubMed] [Google Scholar]
- Bzymek M., Lovett S.T. Instability of repetitive DNA sequences: the role of replication in multiple mechanisms. Proc. Natl. Acad. Sci. U.S.A. 2001;98:8319–8325. doi: 10.1073/pnas.111008398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell I.D., Humphries M.J. Integrin Structure, Activation, and Interactions. Cold Spring Harb Perspect Biol. 2011;3 doi: 10.1101/cshperspect.a004994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chothia C., Hubbard T., Brenner S., Barns H., Murzin A. PROTEIN FOLDS IN THE ALL-β AND ALL-α CLASSES. Annu. Rev. Biophys. 1997;26:597–627. doi: 10.1146/annurev.biophys.26.1.597. [DOI] [PubMed] [Google Scholar]
- Chothia C., Janin J. Relative orientation of close-packed β-pleated sheets in proteins. Proc. Natl. Acad. Sci. U.S.A. 1981;78:4146–4150. doi: 10.1073/pnas.78.7.4146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirillo S.L.G., Bermudez L.E., El-Etr S.H., Duhamel G.E., Cirillo J.D. Legionella pneumophila Entry GenertxA Is Involved in Virulence. Infect. Immun. 2001;69:508–517. doi: 10.1128/IAI.69.1.508-517.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen F.E., Sternberg M.J.E., Taylor W.R. Analysis of the tertiary structure of protein β-sheet sandwiches. J. Mol. Bio. 1981;148:253–272. doi: 10.1016/0022-2836(81)90538-6. [DOI] [PubMed] [Google Scholar]
- Cowtan K. The Buccaneer software for automated model building. 1. Tracing protein chains. Acta Crystallogr D. 2006;62:1002–1011. doi: 10.1107/S0907444906022116. [DOI] [PubMed] [Google Scholar]
- Cucarella C., Solano C., Valle J., Amorena B., Lasa Í., Penadés J.R. Bap, a Staphylococcus aureus Surface Protein Involved in Biofilm Formation. J. Bacteriol. 2001;183:2888–2896. doi: 10.1128/JB.183.9.2888-2896.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Auria G., Jiménez N., Peris-Bondia F., Pelaz C., Latorre A., Moya A. Virulence factor rtx in Legionella pneumophila, evidence suggesting it is a modular multifunctional protein. BMC Genomics. 2008;9:14. doi: 10.1186/1471-2164-9-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delepelaire P., Wandersman C. The SecB chaperone is involved in the secretion of the Serratia marcescens HasA protein through an ABC transporter. EMBO J. 1998;17:936–944. doi: 10.1093/emboj/17.4.936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Y., Li S., Zhao D., Liu J., Ma S., Geng J., Lu C., Liu Y. IolR, a negative regulator of the myo -inositol metabolic pathway, inhibits cell autoaggregation and biofilm formation by downregulating RpmA in Aeromonas hydrophila. npj Biofilms and Microbiomes. 2020;6:1–12. doi: 10.1038/s41522-020-0132-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P., Lohkamp B., Scott W.G., Cowtan K. Features and development of Coot. Acta Crystallogr. D. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans P. Scaling and assessment of data quality. Acta Crystallogr. D. 2006;62:72–82. doi: 10.1107/S0907444905036693. [DOI] [PubMed] [Google Scholar]
- Gerlach R.G., Cláudio N., Rohde M., Jäckel D., Wagner C., Hensel M. Cooperation of Salmonella pathogenicity islands 1 and 4 is required to breach epithelial barriers. Cell. Microbiol. 2008;10:2364–2376. doi: 10.1111/j.1462-5822.2008.01218.x. [DOI] [PubMed] [Google Scholar]
- Griessl M.H., Schmid B., Kassler K., Braunsmann C., Ritter R., Barlag B., Stierhof Y.-D., Sturm K.U., Danzer C., Wagner C., Schäffer T.E., Sticht H., Hensel M., Muller Y.A. Structural Insight into the Giant Ca2+-Binding Adhesin SiiE: Implications for the Adhesion of Salmonella enterica to Polarized Epithelial Cells. Structure. 2013;21:741–752. doi: 10.1016/j.str.2013.02.020. [DOI] [PubMed] [Google Scholar]
- Guo S., Campbell R., Davies P.L., Allingham J.S. Phasing with calcium at home. Acta Cryst. F. 2019;75:377–384. doi: 10.1107/S2053230X19004151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo S., Garnham C.P., Partha S.K., Campbell R.L., Allingham J.S., Davies P.L. Role of Ca2+ in folding the tandem β-sandwich extender domains of a bacterial ice-binding adhesin. FEBS J. 2013;280:5919–5932. doi: 10.1111/febs.12518. [DOI] [PubMed] [Google Scholar]
- Guo S., Garnham C.P., Whitney J.C., Graham L.A., Davies P.L. Re-Evaluation of a Bacterial Antifreeze Protein as an Adhesin with Ice-Binding Activity. PLOS ONE. 2012;7 doi: 10.1371/journal.pone.0048805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo S., Stevens C.A., Vance T.D.R., Olijve L.L.C., Graham L.A., Campbell R.L., Yazdi S.R., Escobedo C., Bar-Dolev M., Yashunsky V., Braslavsky I., Langelaan D.N., Smith S.P., Allingham J.S., Voets I.K., Davies P.L. Structure of a 1.5-MDa adhesin that binds its Antarctic bacterium to diatoms and ice. Sci. Adv. 2017;3 doi: 10.1126/sciadv.1701440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo S., Vance T.D.R., Stevens C.A., Voets I., Davies P.L. RTX Adhesins are Key Bacterial Surface Megaproteins in the Formation of Biofilms. Trends Microbiol. 2019 doi: 10.1016/j.tim.2018.12.003. [DOI] [PubMed] [Google Scholar]
- Hakim A., Nguyen J.B., Basu K., Zhu D.F., Thakral D., Davies P.L., Isaacs F.J., Modis Y., Meng W. Crystal structure of an insect antifreeze protein and its implications for ice binding. J. Biol. Chem. 2013;288:12295–12304. doi: 10.1074/jbc.M113.450973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinsa S.M., Espinosa-Urgel M., Ramos J.L., O’Toole G.A. Transition from reversible to irreversible attachment during biofilm formation by Pseudomonas fluorescens WCS365 requires an ABC transporter and a large secreted protein. Mol. Microbiol. 2003;49:905–918. doi: 10.1046/j.1365-2958.2003.03615.x. [DOI] [PubMed] [Google Scholar]
- Hospenthal M.K., Zyla D., Costa T.R.D., Redzej A., Giese C., Lillington J., Glockshuber R., Waksman G. The Cryoelectron Microscopy Structure of the Type 1 Chaperone-Usher Pilus Rod. Structure. 2017;25:1829–1838.e4. doi: 10.1016/j.str.2017.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov I.E., Boyd C.D., Newell P.D., Schwartz M.E., Turnbull L., Johnson M.S., Whitchurch C.B., O’Toole G.A., Camesano T.A. Atomic force and super-resolution microscopy support a role for LapA as a cell-surface biofilm adhesin of Pseudomonas fluorescens. Res. Microbiol. 2012;163:685–691. doi: 10.1016/j.resmic.2012.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W. Integration, scaling, space-group assignment and post-refinement. Acta Crystallogr D. 2010;66:133–144. doi: 10.1107/S0907444909047374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi T., Mizunoe Y., Takade A., Naito S., Yoshida S. Curli fibers are required for development of biofilm architecture in Escherichia coli K-12 and enhance bacterial adherence to human uroepithelial cells. Microbiol. Immunol. 2005;49:875–884. doi: 10.1111/j.1348-0421.2005.tb03678.x. [DOI] [PubMed] [Google Scholar]
- Kim S.A., Tai C.-Y., Mok L.-P., Mosser E.A., Schuman E.M. Calcium-dependent dynamics of cadherin interactions at cell–cell junctions. Proc. Natl. Acad. Sci. 2011;108:9857–9862. doi: 10.1073/pnas.1019003108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kister A.E., Finkelstein A.V., Gelfand I.M. Common features in structures and sequences of sandwich-like proteins. Proc. Natl. Acad. Sci. 2002;99:14137–14141. doi: 10.1073/pnas.212511499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klemm P., Schembri M.A. Bacterial adhesins: function and structure. Int. J. Med. Microbiol. 2000;290:27–35. doi: 10.1016/S1438-4221(00)80102-2. [DOI] [PubMed] [Google Scholar]
- Koch A.W., Pokutta S., Lustig A., Engel J. Calcium Binding and Homoassociation of E-cadherin Domains. Biochemistry. 1997;36:7697–7705. doi: 10.1021/bi9705624. [DOI] [PubMed] [Google Scholar]
- Leahy D.J., Hendrickson W.A., Aukhil I., Erickson H.P. Structure of a fibronectin type III domain from tenascin phased by MAD analysis of the selenomethionyl protein. Science. 1992;258:987–991. doi: 10.1126/science.1279805. [DOI] [PubMed] [Google Scholar]
- Leo J.C., Elovaara H., Bihan D., Pugh N., Kilpinen S.K., Raynal N., Skurnik M., Farndale R.W., Goldman A. First analysis of a bacterial collagen-binding protein with collagen Toolkits: promiscuous binding of YadA to collagens may explain how YadA interferes with host processes. Infect. Immun. 2010;78:3226–3236. doi: 10.1128/IAI.01057-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Linke W.A., Oberhauser A.F., Carrion-Vazquez M., Kerkvliet J.G., Lu H., Marszalek P.E., Fernandez J.M. Reverse engineering of the giant muscle protein titin. Nature. 2002;418:998–1002. doi: 10.1038/nature00938. [DOI] [PubMed] [Google Scholar]
- Liebschner D., Afonine P.V., Baker M.L., Bunkóczi G., Chen V.B., Croll T.I., Hintze B., Hung L.W., Jain S., McCoy A.J., Moriarty N.W., Oeffner R.D., Poon B.K., Prisant M.G., Read R.J., Richardson J.S., Richardson D.C., Sammito M.D., Sobolev O.V., Stockwell D.H., Terwilliger T.C., Urzhumtsev A.G., Videau L.L., Williams C.J., Adams P.D. Macromolecular structure determination using X-rays, neutrons and electrons: recent developments in Phenix. Acta Crystallogr D Struct Biol. 2019;75:861–877. doi: 10.1107/S2059798319011471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H., Isralewitz B., Krammer A., Vogel V., Schulten K. Unfolding of titin immunoglobulin domains by steered molecular dynamics simulation. Biophys. J. 1998;75:662–671. doi: 10.1016/S0006-3495(98)77556-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Gil M., Yousef-Coronado F., Espinosa-Urgel M. LapF, the second largest Pseudomonas putida protein, contributes to plant root colonization and determines biofilm architecture. Mol. Microbiol. 2010;77:549–561. doi: 10.1111/j.1365-2958.2010.07249.x. [DOI] [PubMed] [Google Scholar]
- Mayor U., Johnson C.M., Daggett V., Fersht A.R. Protein folding and unfolding in microseconds to nanoseconds by experiment and simulation. Proc. Natl. Acad. Sci. 2000;97:13518–13522. doi: 10.1073/pnas.250473497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C., Read R.J. Phaser crystallographic software. J Appl Cryst, J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oude Vrielink A.S., Vance T.D.R., de Jong A.M., Davies P.L., Voets I.K. Unusually high mechanical stability of bacterial adhesin extender domains having calcium clamps. PLOS ONE. 2017;12 doi: 10.1371/journal.pone.0174682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palumbo R.N., Wang C. Bacterial invasin: structure, function, and implication for targeted oral gene delivery. Curr Drug Deliv. 2006;3:47–53. doi: 10.2174/156720106775197475. [DOI] [PubMed] [Google Scholar]
- Peters B., Stein J., Klingl S., Sander N., Sandmann A., Taccardi N., Sticht H., Gerlach R.G., Muller Y.A., Hensel M. Structural and functional dissection reveals distinct roles of Ca2+-binding sites in the giant adhesin SiiE of Salmonella enterica. PLOS Pathogens. 2017;13 doi: 10.1371/journal.ppat.1006418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satchell K.J.F. Structure and function of MARTX toxins and other large repetitive RTX proteins. Annu. Rev. Microbiol. 2011;65:71–90. doi: 10.1146/annurev-micro-090110-102943. [DOI] [PubMed] [Google Scholar]
- Sauer F.G., Remaut H., Hultgren S.J., Waksman G. Fiber assembly by the chaperone-usher pathway. Biochim. Biophys. Acta. 2004;1694:259–267. doi: 10.1016/j.bbamcr.2004.02.010. [DOI] [PubMed] [Google Scholar]
- Schilling J.D., Mulvey M.A., Hultgren S.J. Structure and Function of Escherichia coli Type 1 Pili: New Insight into the Pathogenesis of Urinary Tract Infections. J Infect Dis. 2001;183:S36–S40. doi: 10.1086/318855. [DOI] [PubMed] [Google Scholar]
- Shapiro L., Kwong P.D., Fannon A.M., Colman D.R., Hendrickson W.A. Considerations on the folding topology and evolutionary origin of cadherin domains. Proc. Natl. Acad. Sci. U.S.A. 1995;92:6793–6797. doi: 10.1073/pnas.92.15.6793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh P., Carraher C., Schwarzbauer J.E. Assembly of fibronectin extracellular matrix. Annu. Rev. Cell Dev. Biol. 2010;26:397–419. doi: 10.1146/annurev-cellbio-100109-104020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syed K.A., Beyhan S., Correa N., Queen J., Liu J., Peng F., Satchell K.J.F., Yildiz F., Klose K.E. The Vibrio cholerae Flagellar Regulatory Hierarchy Controls Expression of Virulence Factors. J. Bacteriol. 2009;191:6555–6570. doi: 10.1128/JB.00949-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeichi M. Cadherins: a molecular family important in selective cell-cell adhesion. Annu. Rev. Biochem. 1990;59:237–252. doi: 10.1146/annurev.bi.59.070190.001321. [DOI] [PubMed] [Google Scholar]
- Terwilliger T.C., Adams P.D., Read R.J., McCoy A.J., Moriarty N.W., Grosse-Kunstleve R.W., Afonine P.V., Zwart P.H., Hung L.W. Decision-making in structure solution using Bayesian estimates of map quality: the PHENIX AutoSol wizard. Acta Crystallogr. D. 2009;65:582–601. doi: 10.1107/S0907444909012098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger T.C., Grosse-Kunstleve R.W., Afonine P.V., Moriarty N.W., Zwart P.H., Hung L.-W., Read R.J., Adams P.D. Iterative model building, structure refinement and density modification with the PHENIX AutoBuild wizard. Acta Crystallogr Sect D. 2008;64:61–69. doi: 10.1107/S090744490705024X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai J.C., Yen M.-R., Castillo R., Leyton D.L., Henderson I.R., Saier M.H. The Bacterial Intimins and Invasins: A Large and Novel Family of Secreted Proteins. PLOS One. 2010;5 doi: 10.1371/journal.pone.0014403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagin A.A., Steiner R.A., Lebedev A.A., Potterton L., McNicholas S., Long F., Murshudov G.N. REFMAC5 dictionary: organization of prior chemical knowledge and guidelines for its use. Acta Crystallogr Sect D. 2004;60:2184–2195. doi: 10.1107/S0907444904023510. [DOI] [PubMed] [Google Scholar]
- Valbuena A., Oroz J., Hervás R., Vera A.M., Rodríguez D., Menéndez M., Sulkowska J.I., Cieplak M., Carrión-Vázquez M. On the remarkable mechanostability of scaffoldins and the mechanical clamp motif. Proc. Natl. Acad. Sci. U.S.A. 2009;106:13791–13796. doi: 10.1073/pnas.0813093106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vance T.D.R., Guo S., Assaie-Ardakany S., Conroy B., Davies P.L. Structure and functional analysis of a bacterial adhesin sugar-binding domain. PLOS ONE. 2019;14 doi: 10.1371/journal.pone.0220045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vance T.D.R., Olijve L.L.C., Campbell R.L., Voets I.K., Davies P.L., Guo S. Ca2+-stabilized adhesin helps an Antarctic bacterium reach out and bind ice. Biosci Rep. 2014;34 doi: 10.1042/BSR20140083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner C., Barlag B., Gerlach R.G., Deiwick J., Hensel M. The Salmonella enterica giant adhesin SiiE binds to polarized epithelial cells in a lectin-like manner. Cell. Microbiol. 2014;16:962–975. doi: 10.1111/cmi.12253. [DOI] [PubMed] [Google Scholar]
- Williams A.F., Barclay A.N. The Immunoglobulin Superfamily—Domains for Cell Surface Recognition. Annu. Rev. Immun. 1988;6:381–405. doi: 10.1146/annurev.iy.06.040188.002121. [DOI] [PubMed] [Google Scholar]
- Winn M.D., Ballard C.C., Cowtan K.D., Dodson E.J., Emsley P., Evans P.R., Keegan R.M., Krissinel E.B., Leslie A.G.W., McCoy A., McNicholas S.J., Murshudov G.N., Pannu N.S., Potterton E.A., Powell H.R., Read R.J., Vagin A., Wilson K.S. Overview of the CCP4 suite and current developments. Acta Crystallogr Sect D. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrobel, A., Ottoni, C., Leo, J.C., Gulla, S., Linke, D., 2018. The repeat structure of two paralogous genes, Yersinia ruckeri invasin (yrInv) and a “Y. ruckeri invasin-like molecule”, (yrIlm) sheds light on the evolution of adhesive capacities of a fish pathogen. J. Struct. Biol., Protein with Tandem Repeats: Sequences, Structures and Functions 201, 171–183. https://doi.org/10.1016/j.jsb.2017.08.008. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
X-ray crystal structure coordinates solved in this study have been deposited in the Protein Data Bank with accession codes 6XI3 (MhLap) and 6XI1 (AhLap). Data that support the findings of this study are available from the corresponding author P.L.D. upon reasonable request.