Abstract
Background:
Disabled homolog 2-interacting protein is a new member of the Ras GTPase superfamily involved in the regulation of cell proliferation, apoptosis, and metastasis. However, the expression of disabled homolog 2-interacting protein in renal cell carcinoma, its correlation with cancer prognosis, and tumor infiltrating lymphocytes remains unclear.
Methods:
The expression of disabled homolog 2-interacting protein was analyzed by UALCAN database, GEPIA database and the evaluation of disabled homolog 2-interacting protein effects on clinical prognosis. Prognostic factor analysis was used to identify the correlations between disabled homolog 2-interacting protein and cancer immune infiltration via the TIMER database. In addition, COXPRESdb database was used to analyze the enrichment of disabled homolog 2-interacting protein co-expression genes.
Results:
Compared to the normal tissues, the messenger RNA expression levels of DAB2IP are higher in 8 while lower in 15 types of tumor tissues. Furthermore, disabled homolog 2-interacting protein has high expression in kidney chromophobe and low expression in both kidney renal clear cell carcinoma and kidney renal papillary cell carcinoma. The messenger RNA expression levels of disabled homolog 2-interacting protein decrease gradually due to the increasing tumor staging which positively correlates with disease-free survival and overall survival in both kidney renal clear cell carcinoma and kidney renal papillary cell carcinoma. The expression levels of disabled homolog 2-interacting protein also positively correlate with the tumor purity of kidney chromophobe, kidney renal clear cell carcinoma, and kidney renal papillary cell carcinoma samples. Besides, the expression of disabled homolog 2-interacting protein in renal cell carcinoma has negative correlation with the immune infiltration, and the immune infiltration of B cells and CD8+ T cells affects the prognosis of kidney renal papillary cell carcinoma. Enrichment analysis of disabled homolog 2-interacting protein co-expressed genes suggested that its biological role was mainly in regulating GTPase activity.
Conclusions:
These findings suggest that disabled homolog 2-interacting protein functions as a tumor suppressor in the progression of renal cell carcinoma, and the expression of disabled homolog 2-interacting protein is related to the immune infiltrating cells and affects the survival of renal cell carcinoma. Disabled homolog 2-interacting protein can be a novel clinical biomarker for patients with renal cell carcinoma, which also provides new insights for the future treatments of renal cell carcinoma.
Keywords: DAB2IP, renal cell carcinoma, mRNA, biomarker, prognosis, immune infiltration
Introduction
There were 73 750 cases of renal carcinoma and renal pelvis carcinoma in 2020 in the United States according to statistics, accounting for 5% and 3% of the new cancer cases in males and females, respectively.1 Renal cell carcinoma (RCC) is essentially the most common solid tumor of the kidney and the most deadly malignancy of the urinary system.2 As a heterogeneous disease, kidney renal clear cell carcinoma (KIRC), kidney renal papillary cell carcinoma (KIRP), and kidney chromophobe (KICH) are the most common pathological subtypes of RCC.3,4 In recent years, more and more scholars believe that different subtypes of RCC have different histology, genetic alterations, clinical characteristics, and prognosis.5-7 In cases of RCC with only local invasion, surgery remains the primary treatment and has a good long-term survival rate. Once metastasis occurs, the tumor is difficult to treat because traditional chemotherapy and radiotherapy are not effective in treating RCC.8 With the advent of targeted therapies, although the treatment of advanced RCC has changed significantly, patient survival has generally increased by months due to acquired resistance to these therapies. Therefore, further research is needed to explore the biomarkers for the early diagnosis of kidney cancer as well as a potential therapeutic target for RCC.
Human disabled homolog 2-interacting protein (DAB2IP) was first reported in 2002 as a protein-coding gene located on the human 9q33.1-q33.3 chromosome.9 Disabled homolog 2-interacting protein is a tumor suppressor gene associated with ovarian cancer,10 prostate cancer,11,12 breast cancer,13 and choriocarcinoma.14 In recent years, increasing studies have found that DAB2IP is significantly downregulated in many types of cancer, and the low expression of DAB2IP in tumors may predict the poor prognosis of malignant tumors.15-17
Previous studies have shown that downregulated DAB2IP expression can produce radiation-resistant properties in RCC18 and mTOR-targeted therapy resistance19 and suggested that the low expression of DAB2IP may be an important factor for poor prognosis of RCC. However, specific levels of DAB2IP expression in various pathological subtypes of RCC are still lacking, and the potential function and mechanism of DAB2IP in tumor immunology are rarely discussed. In this study, we use multidimensional analysis strategy to investigate the expression, prognosis, and immune infiltration of DAB2IP in RCC reported on different online databases. Our findings shed light on the important role of DAB2IP in RCC which investigates the correlation between the levels of DAB2IP expression and tumor immune cell infiltration and provides a future direction for the development of new immune therapeutic target in RCC.
Materials and Methods
Correlation Analysis of Gene Expression in UALCAN Database
UALCAN (http://ualcan.path.uab.edu/index.html) is a user-friendly, interactive website for analyzing cancer transcriptome data.20 UALCAN contains published cancer transcriptome data (The Cancer Genome Atlas [TCGA] and MET500 transcriptome sequencing). By using the UALCAN database, we determined the levels of DAB2IP gene expression in various types of cancer and further revealed the levels of DAB2IP expression in the 3 most common pathological subtypes of RCC. When P < .05, the difference was considered statistically significant.
Correlation Analysis of Gene Expression in Gene Expression Profile Interactive Analysis
The Gene Expression Profile Interactive Analysis (GEPIA) (http://gepia.cancer-pku.cn/index.html) online database is used to analyze the relationship between DAB2IP expression and tumor stage in RCC. GEPIA21 is an interactive website that covers 9736 tumors and 8587 normal tissue samples from TCGA and GTEx databases and analyzes the results of RNA sequencing expression. GEPIA’s log-rank test and Mantel-Cox test based on the expression of DAB2IP generated kidney cancer survival curves, including disease-free survival (DFS) and overall survival (OS). Log-rank P value of <.05 was considered statistically significant for survival between different groups, and P (hazard ratio [HR]) value of <.05 was considered statistically significant for the HR.
The TIMER Analysis
TIMER is a comprehensive resource for systematic analysis of immune information for various tumor types (https://cistrome.shinyapps.io/timer/).22 TIMER used previously published deconvolution statistics23 to infer the abundance of tumor-infiltrated immune cells (including B cells, CD8+ T cells, CD4+ T cells, and macrophages, neutrophils, and dendritic cells) from the gene expression profiles. We analyzed the correlation between the expression of DAB2IP and the infiltration level of different immune cells in the 3 pathological subtypes of RCC through the GENE module. Kaplan-Meier curves of immune infiltrates were drawn via SURVIVAL module, and the survival difference curve of RCC was drawn according to different levels of DAB2IP expression. Log-rank P < .05 was considered statistically significant. We further used the SCAN module in the TIMER platform to analyze the relationship between infiltration level and DAB2IP mutations in RCC.
Co-Expression Analysis in COXPRESdb
COXPRESdb (https://coxpresdb.jp) is a database providing co-expression information for 11 species of animals.24 One of its major features is to compare the multiple co-expression data from different transcriptomics techniques and different species. We searched the top 50 co-expressed genes that most relevant to DAB2IP in kidney cancer on the COXPRESdb website as well as the DAB2IP and the selected 50 genes to complete the next study.
Function Enrichment Analysis by R Software
Functional enrichment analysis classifies gene lists based on gene function and its correlation with biological phenotypes. We performed GO and KEGG analysis on DAB2IP and the above 50 co-expressed genes through clusterProfiler25 package in R software to understand the biological process of the genome, cellular component, molecular function, and the most important signal pathway. P < .05 was considered statistically significant and included in the analysis.
Results
Expression of DAB2IP on Transcriptional Level in Various Cancers
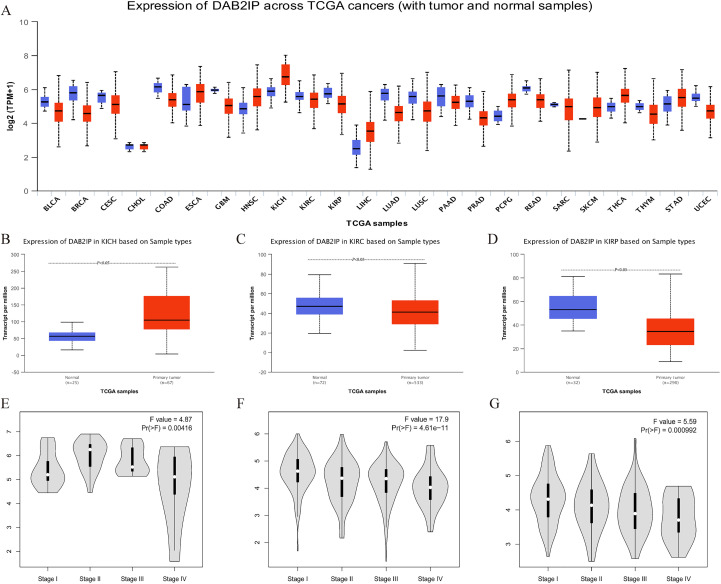
In order to analyze the difference in DAB2IP expression between tumors and normal tissues, we compared the messenger RNA (mRNA) expression levels of DAB2IP between tumors and normal tissues of various cancer types for the TCGA database by using the UALCAN platform (Figure 1A). The results showed that the mRNA expression levels of DAB2IP, compared to correspondingly normal tissue, is significantly higher in 8 types of tumor tissues (ESCA, HNSC, KICH, LIHC, PCPG, SKCM, THCA, and STAD) while significantly lower in 15 types of tumor tissues (BLCA, BRCA, CESC, COAD, GBM, KIRC, KIRP, LUAD, LUSC, PAAD, PRAD, READ, SARC, THYM, and UCEC).
Figure 1.
DAB2IP expression in different cancers, especially in RCC. A, DAB2IP expression on transcriptional level in various cancers (UALCAN). B-D, Relationship between the transcriptional level of DAB2IP and clinical pathology in RCC (UALCAN). E-G, Relationship between the transcriptional level of DAB2IP and tumor stage in RCC (GEPIA). DAB2IP indicates disabled homolog 2-interacting protein; GEPIA, Gene Expression Profile Interactive Analysis; RCC, renal cell carcinoma.
Relationship Between the Transcriptional Level of DAB2IP and Clinical Pathology and Tumor Stage in RCC
To explore the relationship between transcriptional level of DAB2IP and tumor staging of the specific pathology in RCC, we analyzed the levels of DAB2IP expression in KICH, KIRC, and KIRP samples, respectively, via the UALCAN platform (Figure 1B-D). The box plot intuitively reflects that compared to the normal tissues, the mRNA expression of DAB2IP is significantly increased in KICH. While the mRNA expression of DAB2IP is significantly decreased in KIRC and KIRP compared to correspondingly normal tissues (all P < .05). Violin-like plots showed that the mRNA expression level of DAB2IP gradually decreased with the increase of tumor staging in KIRC and KIRP, while there was no linear relationship between mRNA expression of DAB2IP and tumor staging in KICH (Figure 1E-G).
Relationship Between the Transcriptional Level of DAB2IP and Survival Rate for RCC
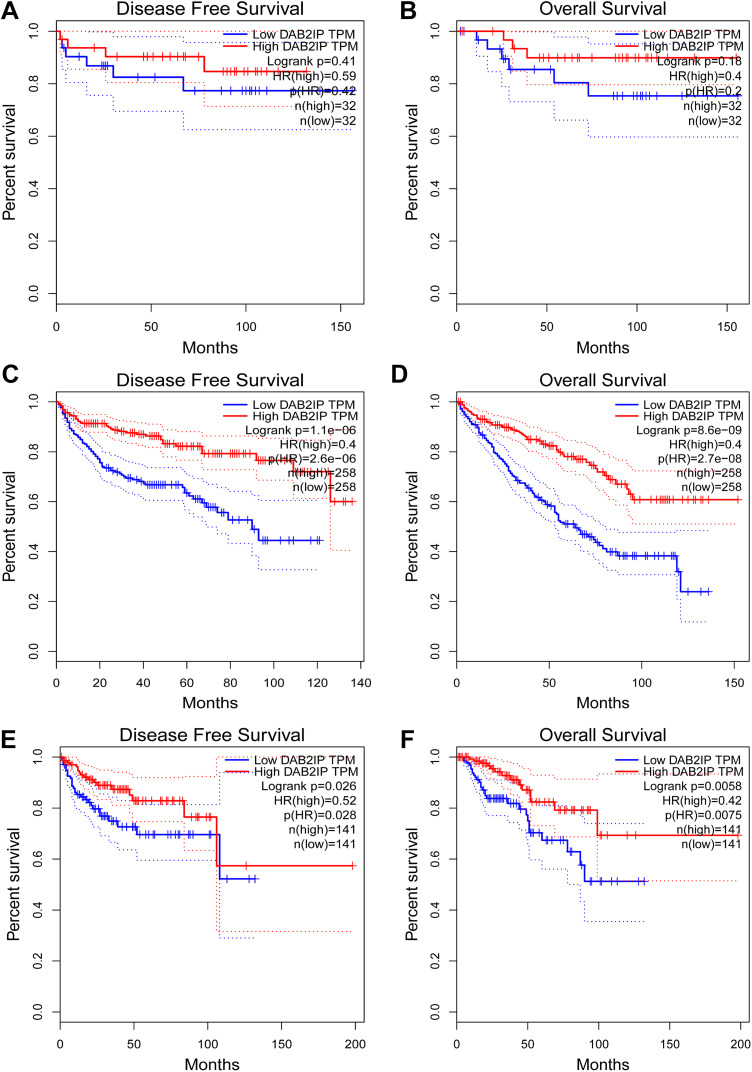
In this study, we continued to explore the correlation between DAB2IP expression and prognosis in RCC. The results showed that higher the DAB2IP expression, better the DFS and OS will be in KICH (respectively, P = .41 and P = .18; Figure 2A and B). In KIRC and KIRP, the mRNA expression of DAB2IP was positively correlated with DFS and OS (all, P < .05; Figure 2C-E).
Figure 2.
Prognostic value of mRNA expression of DAB2IP in RCC (GEPIA). A and B, The relationship between the transcriptional level of DAB2IP and survival rate in KICH. C and D, The relationship between the transcriptional level of DAB2IP and survival rate in KIRC. E and F, The relationship between the transcriptional level of DAB2IP and survival rate in KIRP. DAB2IP indicates disabled homolog 2-interacting protein; RCC, renal cell carcinoma; GEPIA, Gene Expression Profile Interactive Analysis; mRNA, messenger RNA.
Relationship Between DAB2IP Expression and Immune Infiltrates in RCC
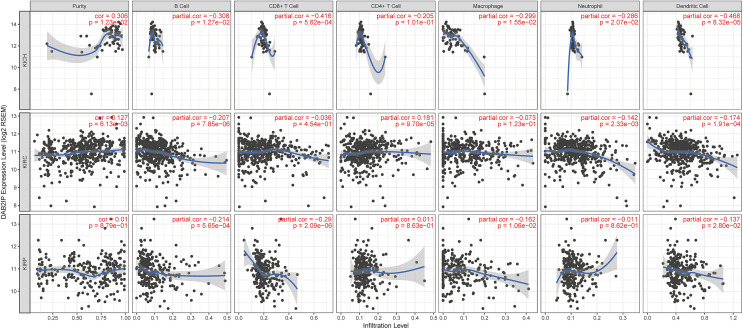
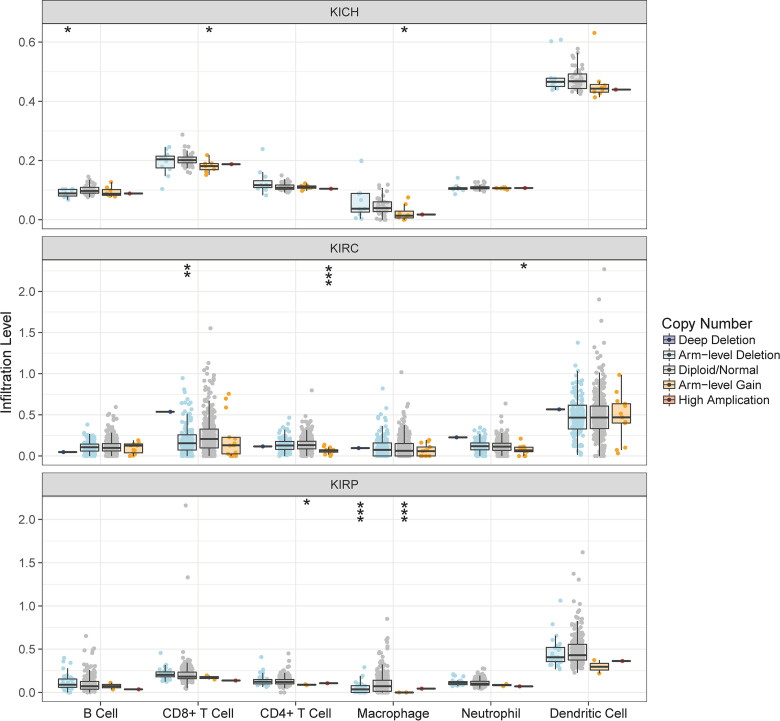
To explore the relationship between the DAB2IP expression and the immune infiltration, we investigated the levels of DAB2IP expression in RCC through the TIMER platform (Figure 3). The results show a positive correlation between the levels of DAB2IP expression with the tumor purity of KICH, KIRC, and KIRP samples (respectively, cor = .306, P = .012; cor = .127, P = .006; cor = .01, P = .879). We also observed a correlation between the expression of DAB2IP and the tumor infiltrating cells. The levels of DAB2IP expression are negatively correlated with the tumor infiltrating cells in KICH (all, P < .05), except CD4+ T cells (P = .101; Figure 3-KICH). In KIRC, the levels of DAB2IP expression are negatively correlated with B cells, neutrophils, and dendritic cells, while positively correlated with CD4+ T cells (all, P < .05; Figure 3-KIRC). In KIRP, the levels of DAB2IP expression are negatively correlated with B cells, CD8+ T cells, macrophages, and dendritic cells (all, P < .05) while positively correlated with CD4+ T cells (P = .863; Figure 3-KIRP).
Figure 3.
The relationship between DAB2IP expression and immune infiltrates in RCC (TIMER). DAB2IP indicates disabled homolog 2-interacting protein; RCC, renal cell carcinoma.
Correlation Between Survival and Abundance of Immune Infiltration in RCC
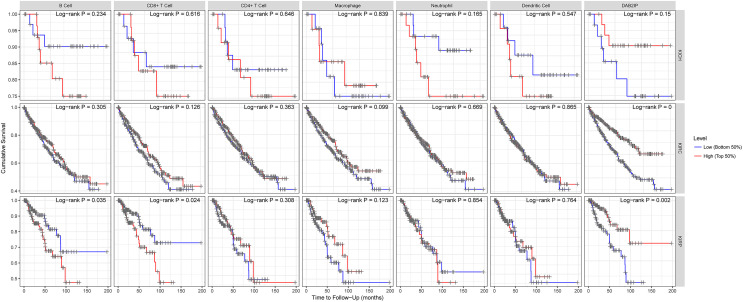
To analyze the relationship between various immune infiltration cell and the survival rate of RCC, we explored the effects of the infiltration immune cell levels on the prognosis of different types in RCC by using the TIMER platform. The relationship between the DAB2IP expression and survival demonstrates the result that the lower level of the various immune-infiltrating cells in KICH, except macrophages, indicates better survival, but the finding lacks statistical significance (Figure 4-KICH). For KIRC, although the result did not reach statistical significance, higher the level of various immune-infiltrating cells indicates better survival, consistently with the relationship between DAB2IP expression and survival (P < .05; Figure 4-KIRC). In contrast to the relationship between DAB2IP expression and survival (P = .002), fewer the B cells and the CD8+ T cells in KIRP, better the survival (respectively, P = .035 and P = .024; Figure 4-KIRP).
Figure 4.
The correlation between survival and abundance of immune infiltration in RCC (TIMER). RCC indicates renal cell carcinoma.
Distribution of the Abundance of Tumor-Infiltrating Immune Cells Under Different Mutation States of the DAB2IP Gene
To analyze the relationship between gene mutations and immune infiltration, we compared the distribution of the abundance of tumor-infiltrating immune cells under different mutation states of the DAB2IP (Figure 5). In KICH, CD8+ T cell and macrophage are the main infiltrating immune cells in the arm-level gain state of DAB2IP gene, while B cell is the main infiltrating immune cells in the arm-level deletion state (all P < .05). In KIRC, CD4+ T cell and neutrophils are the main infiltrating immune cells in the arm-level gain state of DAB2IP gene, while CD8+ T cell is the main infiltrating immune cells in the arm-level deletion state (all P < .05). In KIRP, CD4+ T cell and macrophage are the main infiltrating immune cells in the arm-level gain state of DAB2IP gene, while macrophage is the main infiltrating immune cells in the arm-level deletion state (all P < .05).
Figure 5.
The distribution of the abundance of tumor-infiltrating immune cells under different mutation states of the DAB2IP gene (TIMER). DAB2IP indicates disabled homolog 2-interacting protein.
Functional Enrichment Analysis of DAB2IP-Related Co-Expressed Genes in RCC
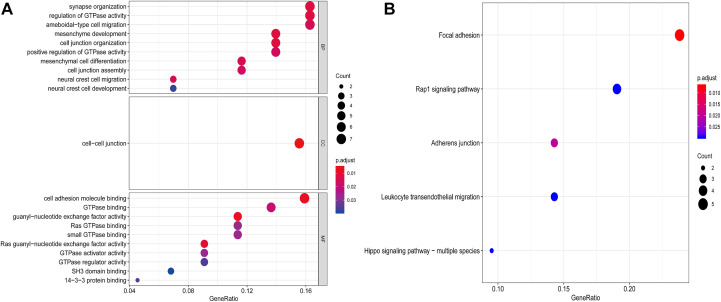
The website COXPRESdb is used to determine the co-expressed gene appear in RCC in relation to the DAB2IP gene, and the top 50 most relevant co-expressed genes are selected (Table 1). Then, the R software is used to generate the GO enrichment analysis and KEGG pathway analysis for the DAB2IP gene and the 50 co-expressed gene mentioned earlier. The results show (Figure 6A) that the biological process mainly revolves around synapse organization, regulation of GTPase activity, and ameboidal-type cell migration; cellular component is mainly enriched at the cell–cell junction; molecular function is mainly involved in cell adhesion molecule binding and GTPase binding; KEGG pathway enrichment analysis shows that focal adhesion and Rap1 signaling pathway are the most important signaling pathways (Figure 6B).
Table 1.
Co-expression genes of DAB2IP via the website COXPRESdb.
| Gene | ||||
|---|---|---|---|---|
| PLEKHG5 | LOC105376257 | CCDC85C | PCDH1 | ARHGAP23 |
| SPTBN2 | SPTBN3 | SPTBN4 | SPTBN5 | SPTBN6 |
| TSPAN9 | BCAR1 | DOCK6 | REXO1 | KAZN |
| LAMA5 | EFNB1 | B4GALNT4 | NECTIN1 | EXD3 |
| SEMA3F | KIAA1217 | RAI1 | HR | KIAA1522 |
| PHLDB1 | ST5 | TNKS1BP1 | AFDN | EPB41L4A-AS2 |
| TEAD3 | PTPRF | CASKIN2 | SPTBN9 | AGAP1 |
| TMEM132A | FBLIM1 | SPTBN8 | LRP5 | ZCCHC14 |
| CARD10 | RBFOX2 | RERE | PPP1R13B | FOXK1 |
| TEAD2 | SPTBN7 | RAPGEF1 | SNCG | MVB12B |
Figure 6.
Functional enrichment analysis of DAB2IP-related co-expressed genes in RCC. A, Co-expression network of DAB2IP is performed by function enrichment analysis by GO in RCC. B, Co-expression network of DAB2IP is performed by function enrichment analysis by KEGG in RCC.
Discussion
Disabled homolog 2-interacting protein is a new member of the Ras GTPase-activating protein family.26 The preferred name of DAB2IP is disabled homolog 2-interacting protein, also called ASK-interacting protein 1, ASK1-interacting protein 1, DAB2 interaction protein, DOC-2/DAB2 interactive protein, and nGAP-like protein. The DAB2IP gene is conserved in chimpanzee, Rhesus monkey, dog, cow, mouse, rat, chicken, zebrafish, and frog, and 258 organisms have orthologs with human gene DAB2IP. A study used RNA-seq in normal tissue displays that DAB2IP has high expressed in colon (RPKM 8.5), testis (RPKM 8.5), kidney (RPKM 7.1), and other tissues.27 In our study, compared to the corresponding normal tissues, DAB2IP has significantly higher expression in KICH while significantly lower expression in KIRC and KIRP. This phenomenon is also consistent with the actual clinical situation. Because the progression and growth of the tumor is slower in KICH, and the survival is better, it is quicker in KIRC and KIRP, and the survival is poorer.2 Consistent with previous studies,9,15,17 this result indicates the tumor suppressive role of DAB2IP in cancer progression. Zhou and colleagues reported that DAB2IP appears to be a new predictive marker for patients with mRCC.19 Wang and colleagues identified 1 CpG methylation biomarker (DAB2IP CpG1) located on UTSS of DAB2IP that was associated with poor overall survival in a cohort of 318 ccRCC.28 In summation, the expression of DAB2IP might be used as a predictor of the diagnosis in KIRC and KIRP.
In this study, we used bioinformatics methods to elucidate the relationship between the level of DAB2IP expression gene and the survival of RCC. On the one hand, the mRNA expression of DAB2IP gradually decreased due to the increased tumor stage in KIRC and KIRP while positively correlated with DFS and OS. Our findings are consistent with a couple of studies including prostate cancer,9 hepatocellular carcinoma,29 gastrointestinal tumor,30 lung cancer,31 breast cancer,13 and bladder cancer.32 On the other hand, tumor cells can use various immune cells to achieve the purpose of proliferation and spread. Therefore, we continued to explore the impact of the multiple immune cell infiltration level on the survival in RCC, and the results indicate lower the levels of B cells and CD8+ T cells in KIRP, better the survival rate will be. This finding is consistent with the study by Yeh et al that reported infiltrating T cells promoting RCC progression.33 Combining the abovementioned 2 results, we have reasons to assume that lower expression of DAB2IP might be an independent risk factor and lead to poor prognosis in patients with KIRC and KIRP.
In this article, we also analyzed the relationship between the levels of DAB2IP expression and prognosis and treatments. Firstly, the level of DAB2IP expression has positive correlation with the tumor purity in RCC, which means the level of DAB2IP expression also has positive correlation with prognosis. Some studies have proved that the higher purity of the tumor, the better the prognosis.34-36 Secondly, tumor infiltrating immune cells and molecules in the microenvironment jointly promote immune escape, growth, and metastasis of tumor.37,38 Our study suggested that fewer the B cells and CD8+ T cells in KIRP, better the survival will be. This finding was confirmed by the study by Yeh et al that pointed out that infiltrating T cells may promote RCC cell invasion via increasing the RCC cell ERβ expression to inhibit the tumor suppressor DAB2IP signals.33 Thirdly, our study found an association between DAB2IP mutations and immune infiltration cells, for example, the arm-level gain or deletion of DAB2IP might be the most statistically significant mutations in immune infiltration cells. A study found loss of DAB2IP in RCC cells enhances their sensitivities to growth factor stimulation and resistances to SMI (such as mammalian target of rapamycin [mTOR] inhibitors).19 Fourthly, function enrichment analysis of DAB2IP co-expressed genes in RCC mainly focuses on synapse organization, regulation of GTPase activity, and GTPase binding in GO, focal adhesion, and Rap1 signaling pathway in KEGG. The study by Zhou suggests that DAB2IP participates in extracellular signal-regulated kinase/RSK1 and phosphoinositide-3 kinase/mTOR pathway.19 Overall, we are convinced that DAB2IP bears an important clinical significance in RCC.
Based on our analysis, we have reasons to believe that DAB2IP acts as a tumor suppressor in the development of kidney cancer, consistent with its role in ovarian, prostate, and mammary cancer as well as choriocarcinoma.26 Furthermore, DAB2IP is frequently downregulated or inactivated in RCC by different mechanisms. For example, DAB2IP mRNA expression was regulated by DNA methylation in vitro.39 Moreover, DAB2IP-related molecules play an important role in the progression of kidney cancer, such as PARP-1, miR-138, DAB2IP CpG1, EZH2, and ABCA13.28,40 Fourthly, infiltrating T cells may promote RCC cell invasion via decreased DAB2IP expression.33 Combining the information mentioned earlier, we can summarize that DAB2IP requires further study in RCC.
In summary, DAB2IP acts as a tumor suppressor in the progression of kidney cancer. DAB2IP emerges to be a novel prognostic/predictive biomarker for patients with RCC, and its function provides a new insight into the molecular mechanisms of drug resistance to mTOR inhibitors, which also can be used to develop new strategies to overcome drug-resistant RCC. Moreover, the expression of DAB2IP is related to the immune infiltrating cells and affects the survival of RCC, which worth further study. At the same time, there are many shortcomings in our research. On the one hand, our results have not been verified in clinical samples and cannot provide accurate clinical data. On the other hand, our research has limit knowledge about the roles of DAB2IP in KICH. The relationship between immune infiltration and prognosis in KIRC needs to be further explored. We will conduct more related research in the future.
Supplemental Material
Supplemental Material, coexpression for DAB2IP Plays Important Clinical Significance and Correlates With Immune Infiltration in Renal Cell Carcinoma by Haoyuan Cao, Jiandong Zhang and Wei Wang in Technology in Cancer Research & Treatment
Abbreviations
- DAB2IP
disabled homolog 2-interacting protein
- DFS
disease-free survival
- GEPIA
Gene Expression Profile Interactive Analysis
- HR
hazard ratio
- mRNA
messenger RNA
- KICH
kidney chromophobe
- KIRC
kidney renal clear cell carcinoma
- KIRP
kidney renal papillary cell carcinoma
- OS
overall survival
- PFA
prognostic factor analysis
- RCC
renal cell carcinoma
- TCGA
The Cancer Genome Atlas
Footnotes
Authors’ Note: Haoyuan Cao and Jiandong Zhang contributed equally to this work. The relevant data involved in this study are from online databases, and there are no new animal and human studies.
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This study was supported by grants from the National Natural Science Foundation of China (81771720).
ORCID iD: Haoyuan Cao  https://orcid.org/0000-0001-7083-6717
https://orcid.org/0000-0001-7083-6717
References
- 1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70(1):7–30. [DOI] [PubMed] [Google Scholar]
- 2. Bhatt JR, Finelli A. Landmarks in the diagnosis and treatment of renal cell carcinoma. Nat Rev Urol. 2014;11(9):517–525. [DOI] [PubMed] [Google Scholar]
- 3. Cohen HT, McGovern FJ. Renal-cell carcinoma. N Engl J Med. 2005;353(23):2477–2490. [DOI] [PubMed] [Google Scholar]
- 4. Amin MB, Amin MB, Tamboli P, et al. Prognostic impact of histologic subtyping of adult renal epithelial neoplasms – an experience of 405 cases. Am J Surg Pathol. 2002;26(3):281–291. [DOI] [PubMed] [Google Scholar]
- 5. Ricketts CJ, De Cubas AA, Fan HH, et al. The cancer genome atlas comprehensive molecular characterization of renal cell carcinoma. Cell Rep. 2018;23(12):3698–3698. [DOI] [PubMed] [Google Scholar]
- 6. Moch H, Cubilla AL, Humphrey PA, Reuter VE, Ulbright TM. The 2016 WHO classification of tumours of the urinary system and male genital organs-part a: renal, penile, and testicular tumours. Eur Urol. 2016;70(1):93–105. [DOI] [PubMed] [Google Scholar]
- 7. Linehan WM, Walther MM, Zbar B. The genetic basis of cancer of the kidney. J Urology. 2003;170(6):2163–2172. [DOI] [PubMed] [Google Scholar]
- 8. Atkins MB, Tannir NM. Current and emerging therapies for first-line treatment of metastatic clear cell renal cell carcinoma. Cancer Treat Rev. 2018;70:127–137. [DOI] [PubMed] [Google Scholar]
- 9. Chen H, Pong RC, Wang Z, Hsieh JT. Differential regulation of the human gene DAB2IP in normal and malignant prostatic epithelia: cloning and characterization. Genomics. 2002;79(4):573–581. [DOI] [PubMed] [Google Scholar]
- 10. Mok SC, Chan WY, Wong KK, et al. DOC-2, a candidate tumor suppressor gene in human epithelial ovarian cancer. Oncogene. 1998;16(18):2381–2387. [DOI] [PubMed] [Google Scholar]
- 11. Zhou J, Scholes J, Hsieh JT. Signal transduction targets in androgen-independent prostate cancer. Cancer Metastasis Rev. 2001;20(3-4):351–362. [DOI] [PubMed] [Google Scholar]
- 12. Tseng CP, Ely BD, Li Y, Pong RC, Hsieh JT. Regulation of rat DOC-2 gene during castration-induced rat ventral prostate degeneration and its growth inhibitory function in human prostatic carcinoma cells. Endocrinology. 1998;139(8):3542–3553. [DOI] [PubMed] [Google Scholar]
- 13. Dote H, Toyooka S, Tsukuda K, et al. Aberrant promoter methylation in human DAB2 interactive protein (hDAB2IP) gene in breast cancer. Clin Cancer Res. 2004;10(6):2082–2089. [DOI] [PubMed] [Google Scholar]
- 14. Fulop V, Colitti CV, Genest D, et al. DOC-2/hDab2, a candidate tumor suppressor gene involved in the development of gestational trophoblastic diseases. Oncogene. 1998;17(4):419–424. [DOI] [PubMed] [Google Scholar]
- 15. Zheng L, Chen K, Zhu L, Su D, Cheng G. Low expression of DAB2IP predicts an unfavorable prognosis in human bladder carcinoma. Onco Targets Ther. 2017;10:5719–5726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Min J, Liu L, Li X, et al. Absence of DAB2IP promotes cancer stem cell like signatures and indicates poor survival outcome in colorectal cancer. Sci Rep. 2015;5:16578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zhang X, Li N, Li X, et al. Low expression of DAB2IP contributes to malignant development and poor prognosis in hepatocellular carcinoma. J Gastroenterol Hepatol. 2012;27(6):1117–1125. [DOI] [PubMed] [Google Scholar]
- 18. Yun EJ, Lin CJ, Dang A, et al. Downregulation of Human DAB2IP gene expression in renal cell carcinoma results in resistance to ionizing radiation. Clin Cancer Res. 2019;25(14):4542–4551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhou J, Luo J, Wu K, et al. Loss of DAB2IP in RCC cells enhances their growth and resistance to mTOR-targeted therapies. Oncogene. 2016;35(35):4663–4674. [DOI] [PubMed] [Google Scholar]
- 20. Chandrashekar DS, Bashel B, Balasubramanya SAH, et al. UALCAN: a portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia. 2017;19(8):649–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45(W1):W98–W102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Li T, Fan J, Wang B, et al. TIMER: a web server for comprehensive analysis of tumor-infiltrating immune cells. Cancer Res. 2017;77(21):e108–e110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Li B, Severson E, Pignon JC, et al. Comprehensive analyses of tumor immunity: implications for cancer immunotherapy. Genome Biol. 2016;17(1):174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Obayashi T, Kagaya Y, Aoki Y, Tadaka S, Kinoshita K. COXPRESdb v7: a gene coexpression database for 11 animal species supported by 23 coexpression platforms for technical evaluation and evolutionary inference. Nucleic Acids Res. 2019;47(D1):D55–D62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16(5):284–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Liu L, Xu C, Hsieh JT, Gong J, Xie D. DAB2IP in cancer. Oncotarget. 2016;7(4):3766–3776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Fagerberg L, Hallstrom BM, Oksvold P, et al. Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Mol Cell Proteomics. 2014;13(2):397–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wang ZR, Wei JH, Zhou JC, et al. Validation of DAB2IP methylation and its relative significance in predicting outcome in renal cell carcinoma. Oncotarget. 2016;7(21):31508–31519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Qiu GH, Xie H, Wheelhouse N, et al. Differential expression of hDAB2IPA and hDAB2IPB in normal tissues and promoter methylation of hDAB2IPA in hepatocellular carcinoma. J Hepatol. 2007;46(4):655–663. [DOI] [PubMed] [Google Scholar]
- 30. Dote H, Toyooka S, Tsukuda K, et al. Aberrant promoter methylation in human DAB2 interactive protein (hDAB2IP) gene in gastrointestinal tumour. Br J Cancer. 2005;92(6):1117–1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yano M, Toyooka S, Tsukuda K, et al. Aberrant promoter methylation of human DAB2 interactive protein (hDAB2IP) gene in lung cancers. Int J Cancer. 2005;113(1):59–66. [DOI] [PubMed] [Google Scholar]
- 32. Shen YJ, Kong ZL, Wan FN, et al. Downregulation of DAB2IP results in cell proliferation and invasion and contributes to unfavorable outcomes in bladder cancer. Cancer Sci. 2014;105(6):704–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Yeh CR, Ou ZY, Xiao GQ, Guancial E, Yeh S. Infiltrating T cells promote renal cell carcinoma (RCC) progression via altering the estrogen receptor beta-DAB2IP signals. Oncotarget. 2015;6(42):44346–44359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mao Y, Feng Q, Zheng P, et al. Low tumor purity is associated with poor prognosis, heavy mutation burden, and intense immune phenotype in colon cancer. Cancer Manag Res. 2018;10:3569–3577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zhang C, Cheng W, Ren X, et al. Tumor purity as an underlying key factor in glioma. Clin Cancer Res. 2017;23(20):6279–6291. [DOI] [PubMed] [Google Scholar]
- 36. Aran D, Sirota M, Butte AJ. Systematic pan-cancer analysis of tumour purity. Nat Commun. 2015;6:8971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Azimi F, Scolyer RA, Rumcheva P, et al. Tumor-infiltrating lymphocyte grade is an independent predictor of sentinel lymph node status and survival in patients with cutaneous melanoma. J Clin Oncol. 2012;30(21):2678–2683. [DOI] [PubMed] [Google Scholar]
- 38. Ohtani H. Focus on TILs: prognostic significance of tumor infiltrating lymphocytes in human colorectal cancer. Cancer Immun. 2007;7:4. [PMC free article] [PubMed] [Google Scholar]
- 39. Lasseigne BN, Brooks JD. The role of DNA methylation in renal cell carcinoma. Mol Diagn Ther. 2018;22(4):431–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Yun EJ, Baek ST, Xie D, et al. DAB2IP regulates cancer stem cell phenotypes through modulating stem cell factor receptor and ZEB1. Oncogene. 2015;34(21):2741–2752. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Material, coexpression for DAB2IP Plays Important Clinical Significance and Correlates With Immune Infiltration in Renal Cell Carcinoma by Haoyuan Cao, Jiandong Zhang and Wei Wang in Technology in Cancer Research & Treatment