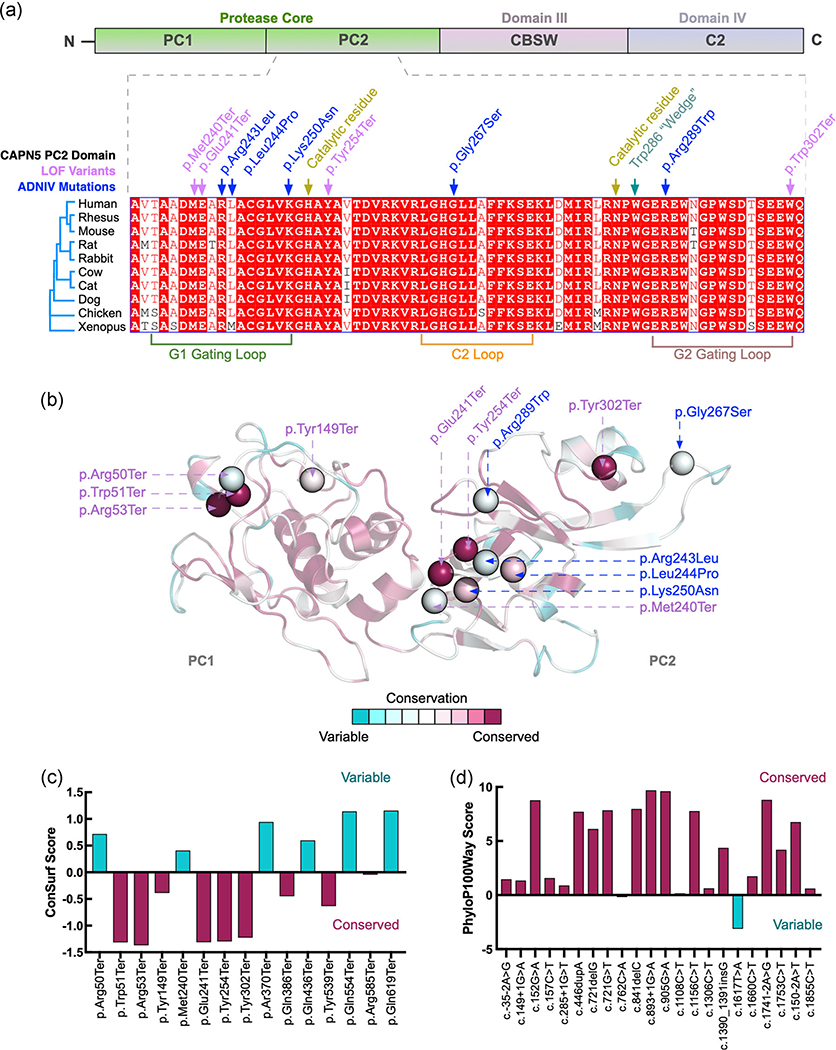
FIGURE 5.
High conservation of variant sites in CAPN5 between invertebrates and vertebrates. (a) Human amino acid sequences from the protease core of CAPN5 (PC2, purple, dashed lines) were aligned to nine specific vertebrates: rhesus, mouse, rat, rabbit, cow, cat, dog, chicken, and xenopus. CAPN5 loss-of-function (LOF) variants (purple) were found in highly conserved regions alongside published autosomal dominant neovascular inflammatory vitreoretinopathy (ADNIV)-disease-causing gain-of-function (GOF) variants (blue). (b) ConSurf conservation analysis using our structural model of CAPN5 as input and comparison to 150 closest homologs including both invertebrates and vertebrates. ConSurf conservation scores were mapped onto our protease core (domain PC2) structural model of CAPN5. (c) Conservation scores were plotted for each of the LOF variants within the protease core (PC2) from 150 analyzed homologs (negative indicates conserved, positive indicates variable amino acid positions). (d) Each of the 22 LOF gene variants for human CAPN5 was used as input into the 100 vertebrate base-wise conservation by PhyloP (phyloP100way). Variants with high conservation, magenta; variants with low conservation, cyan