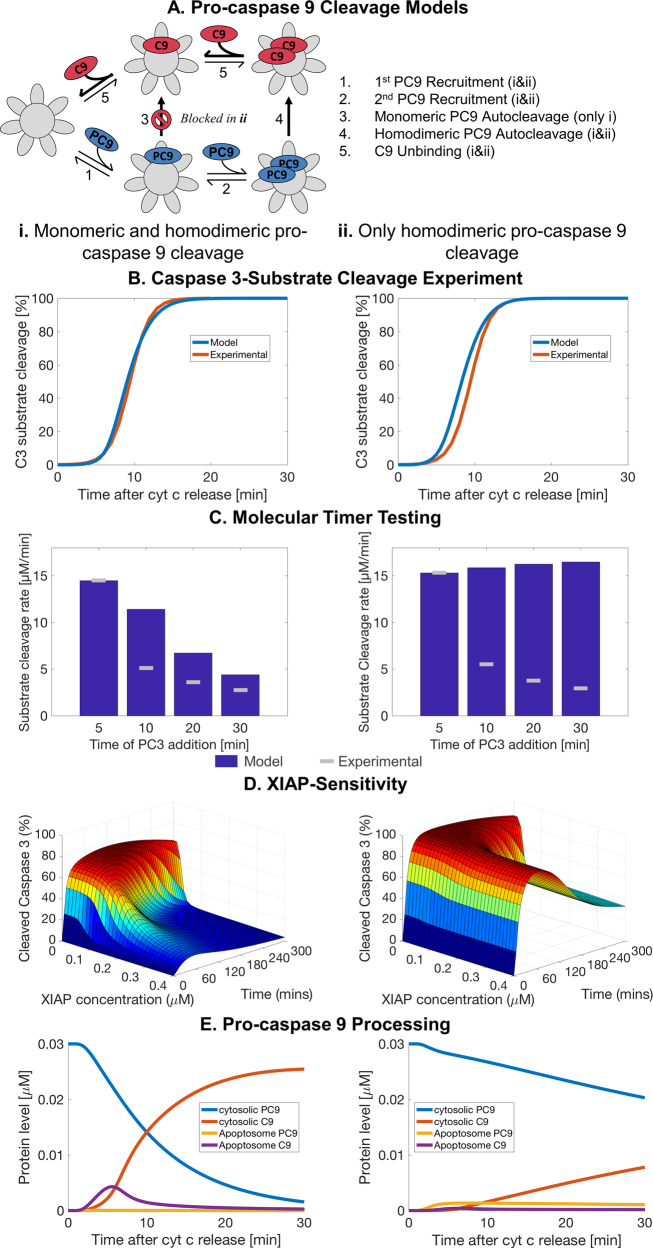
Fig. 1. Mathematical modelling of homodimerisation-mediated autocleavage fails to replicate the apoptosome molecular timer and XIAP-sensitivity.
Comparison of the apoptosis execution models with monomeric PC9 cleavage (i) and with only homodimeric PC9 cleavage (ii) (a). Simulation of C3-substrate cleavage in HeLa cells against experimental data derived from DEVD-FRET probes (b). Simulations of the molecular timer, APAF1 (0.3 μM), PC9 (0.0125 μM), ATP (1 mM) and CytC (10 μM) were incubated for 5–30 min as indicated before addition of PC3 (0.5 μM) and C3-substrate (15 μM) [18]. Values are given as the maximal rate of C3-substrate cleavage (c). Percentage of cleaved C3 (free and XIAP-bound) compared to total PC3+C3 after simulating HeLa cell conditions with different levels of XIAP (d). Percentage of PC9 and C9 over total PC9+C9 in the cytosol and at the apoptosome (e). Experimental data from B sourced from Rehm and colleagues [21]. Experimental data from C sourced from Malladi and colleagues [18].