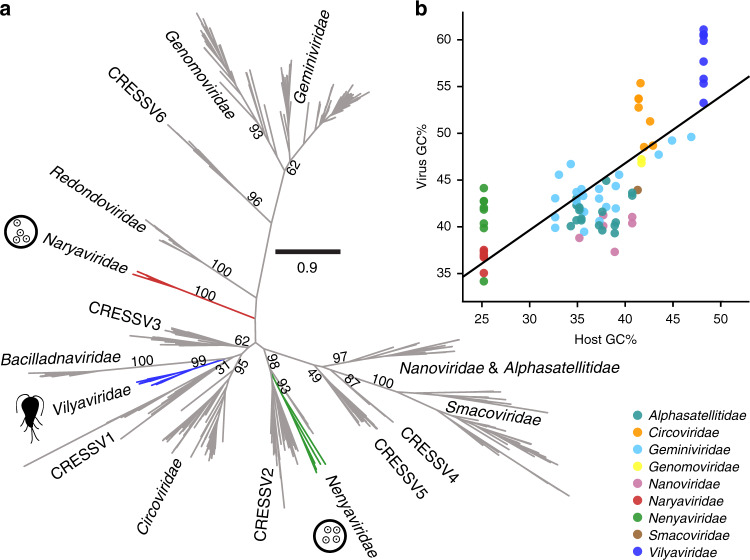
Fig. 2. Parasite-infecting CRESS virus genomes are distinct from known CRESS diversity.
a Phylogenetic maximum-likelihood tree of the Rep protein, scale bar refers to amino acid substitutions per site, numerical values represent bootstrap support of major nodes. The Naryaviridae, Nenyaviridae, and Vilyaviridae contain endogenous viral element sequences extracted from host genomes, respective pictograms of Entamoeba (tetranucleate cyst stage) and Giardia (flagellated trophozoite stage) are shown to indicate this. Five public viral genomes were also found to cluster within these families (MG571899.1, KU043415.1, MH617639.1, KY487991.1 and LC406405.1). b Virus GC-content positively correlates with host GC-content (linear regression, n = 79 biologically independent viral genome sequences, r2 = 0.58, p = 0.01).