Fig. 5.
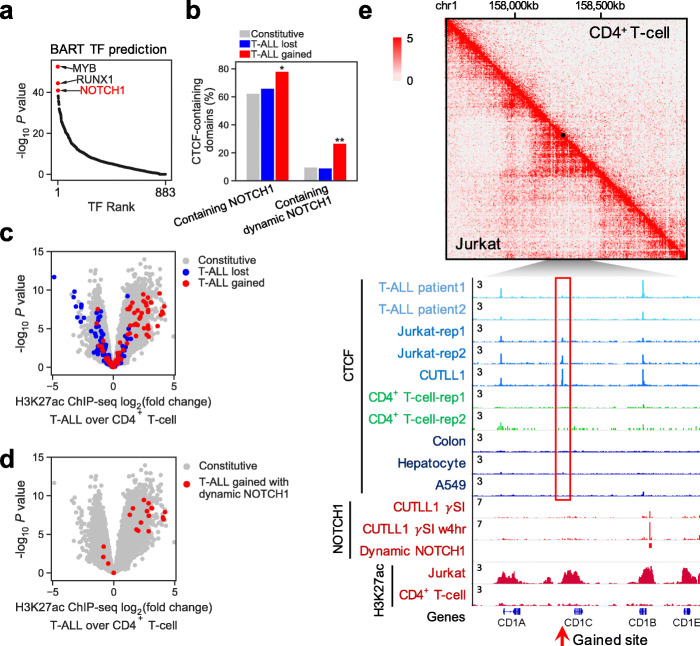
T-ALLgained CTCF binding associates with oncogenic NOTCH1 binding and increased chromatin interaction. a BART-predicted transcription factors binding in genomic regions that have increased interaction with T-ALLgained CTCF sites comparing Jurkat cells with normal CD4+ T cells. b Percentage of chromatin domains including different groups of CTCF binding that contain a NOTCH1 binding site or a dynamic NOTCH1 binding site. *, p < 0.05, **, p < 0.001, by two-tailed Fisher’s exact test. c, d T-ALLgained sites associate with an increased H3K27ac level in Jurkat cells. c Volcano plot showing the differential H3K27ac level between Jurkat cells and normal CD4+ T cells measured by ChIP-seq; each point represents a 10-kb region surrounding a CTCF binding site. d Regions containing dynamic NOTCH1 binding sites are highlighted in red. e Example of Hi-C interaction maps and ChIP-seq tracks around a T-ALLgained CTCF binding site