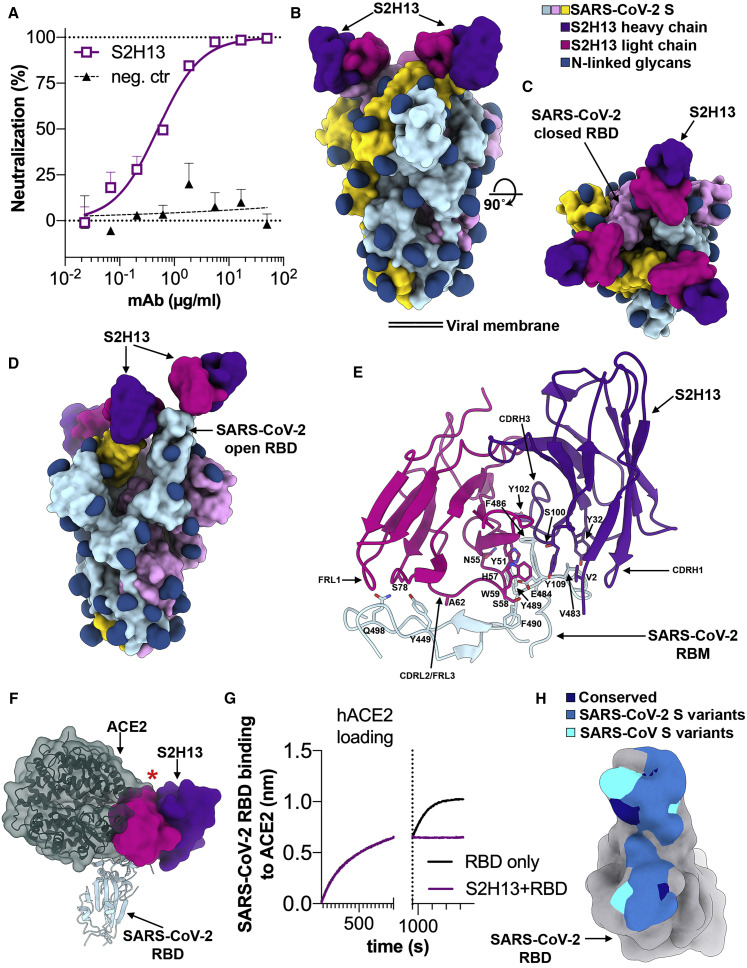
Figure 3.
The S2H13 mAb Inhibits SARS-CoV-2 by Blocking Attachment to ACE2 via Recognition of an Epitope Accessible in the Open and Closed S Conformations
(A) SARS-CoV-2 S pseudovirus neutralization assay indicating an IC50 of 500 ng/mL.
(B and C) Molecular surface representation of the SARS-CoV-2 S/S2H13 Fab complex structure with three RBDs closed shown in two orthogonal orientations.
(D) Molecular surface representation of the SARS-CoV-2 S/S2H13 Fab complex structure with one RBD open. Each SARS-CoV-2 protomer is colored distinctly (cyan, pink, and gold), and N-linked glycans are rendered as dark blue surfaces. The S2H13 light and heavy chain variable domains are colored magenta and purple, respectively.
(E) S2H13 recognizes a crevice formed by the SARS-CoV-2 RBM. Selected side chains at the interface are shown.
(F) S2H13 and ACE2 (dark green) bind overlapping RBM epitope. The red star indicates steric clashes.
(G) BLI binding competition between S2H13 and ACE2 for binding to the SARS-CoV-2 RBD.
(H) Molecular surface representation of the SARS-CoV-2 RBD (gray) with the S2H13 epitope colored by residue conservation across SARS-CoV-2 isolates and SARS-CoV.