Figure 6.
The S304 mAb Promotes SARS-CoV-2 S Opening through Binding to a Cryptic Epitope Conserved within the Sarbecovirus Subgenus
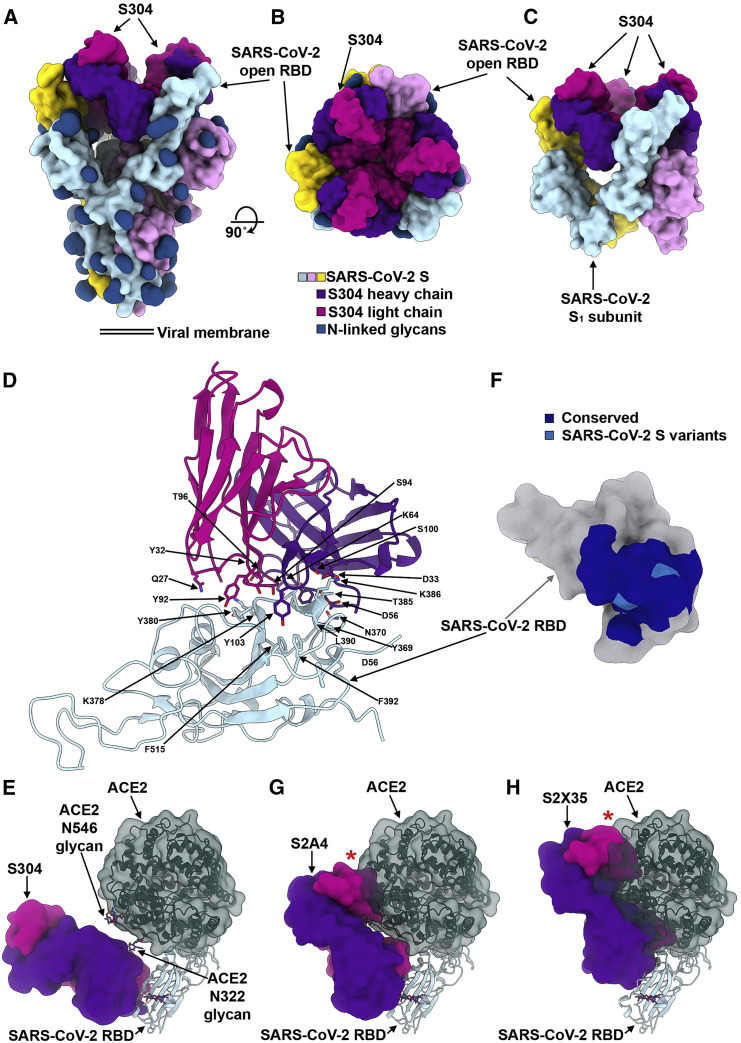
(A and B) Molecular surface representation of the SARS-CoV-2 S/S304 Fab complex cryo-EM structure with three RBDs opened viewed along two orthogonal orientations. Each SARS-CoV-2 S protomer is colored distinctly (cyan, pink, and gold), and N-linked glycans are rendered as dark blue surfaces. The S304 light and heavy chains are colored magenta and purple, respectively.
(C) Cryo-EM reconstruction of the S1 subunit trimer (with disordered S2) bound to three S304 Fabs viewed along two orthogonal orientations and the corresponding atomic model fit in density. Each SARS-CoV-2 S1 protomer is colored distinctly (cyan, pink, and gold). The S304 light and heavy chains are colored magenta and purple, respectively.
(D) Ribbon diagram of the crystal structure of S304 (pink and purple), S2H14, and S309 in complex with the SARS-CoV-2 RBD (light blue). Only the S304 variable domains are shown, whereas S2H14 and S309 were omitted for clarity.
(E) Positioning of ACE2 (dark green) relative to the S304 Fab bound to the SARS-CoV-2 RBD. ACE2 N-linked glycans at position N322 and N546 are indicated, as they could putatively clash with S304.
(F) Molecular surface representation of the SARS-CoV-2 RBD (gray) with the S304 epitope colored by residue conservation with SARS-CoV.
(G and H) Positioning of ACE2 (dark green) relative to the S2A4 (G) and S2X35 (H) Fabs bound to the SARS-CoV-2 RBD. The red stars indicate steric clashes.