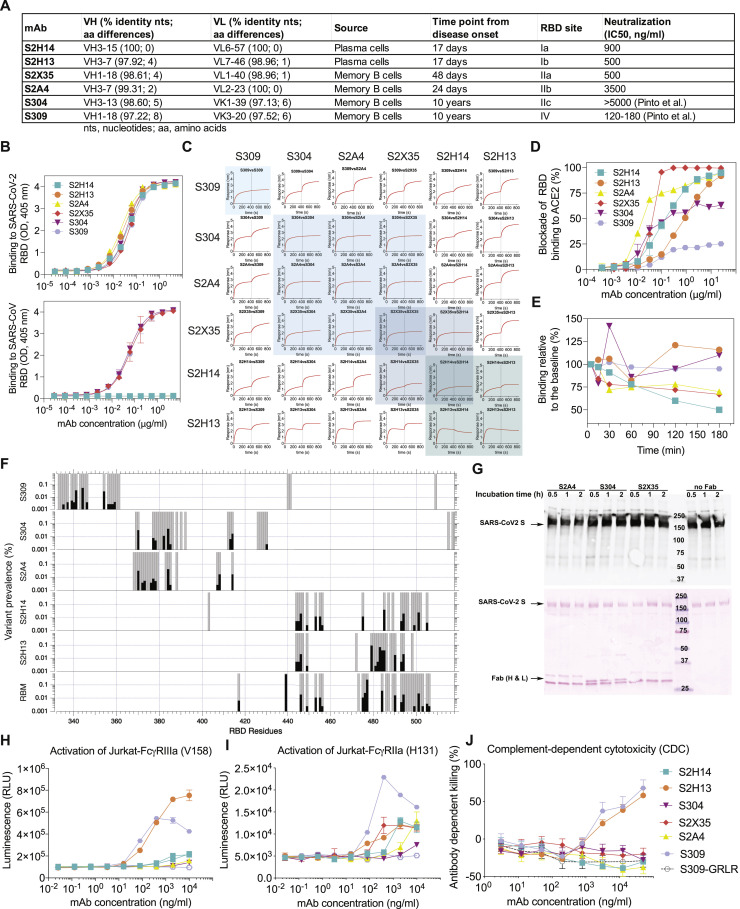
Figure S3.
Characteristics of the Six Probe mAbs Used for Structural and Epitope-Mapping Studies, Related to Figures 3, 4, 5, 6, and 7
(A) V(D)J usage, percentage identity to germline, number of somatic mutations, source and time interval between sample collection and mAb isolation, RBD site recognized and neutralization potency of the 6 mAbs. B mem, memory B cell; PC, plasma cells.
(B) Binding of the 6 mAbs to the SARS-CoV-2 (up) or SARS-CoV (down) RBD analyzed by ELISA.
(C) Competition matrix for binding of each of the six mAbs in presence of another mAb evaluated by biolayer interferometry.
(D) mAb-mediated inhibition of RBD binding to ACE2 analyzed by ELISA.
(E) mAb-mediated S1 subunit shedding from cell-surface expressed SARS-CoV-2 S as determined by flow-cytometry.
(F) Conservation of RBM and epitope residues in ∼74,000 SARS-CoV-2 sequences (GISAID, August 11th, 2020). RBM and epitope residues are shown as gray bars. Black bars indicate variant prevalence for epitope residues with at least 2 variants. RBM residues were determined from PDB 6M0J using a 5.0 Å distance cutoff between RBD and ACE2 residues using MOE.
(G) Western-blot analysis (top) of the prefusion-stabilized SARS-CoV-2 S ectodomain trimer in presence of S2A4, S304 or S2X35 Fab after incubation for the indicated amount of times. Red ponceau staining (bottom) of the SDS-PAGE gel used for carrying out the western blot confirming the presence of added Fabs when indicated.
(H) Analysis of activation of FcγRIIIa (V158 allele) expressed on Jurkat cells by SARS-CoV-2 S stably transfected CHO cells incubated with mAbs. GRLR indicates an antibody Fc variant carrying mutations that abolish binding to FcγRs.
(I) Analysis of activation of FcγRIIa (H131 allele), expressed on Jurkat cells by SARS-CoV-2 S stably transfected CHO cells incubated with mAbs.
(J) Killing of SARS-CoV-2 S stably transfected CHO cells by mAbs in the presence of complement (CDC assay).