Fig. 5. Virus escape through glycan shift revealed by SGA and mutations.
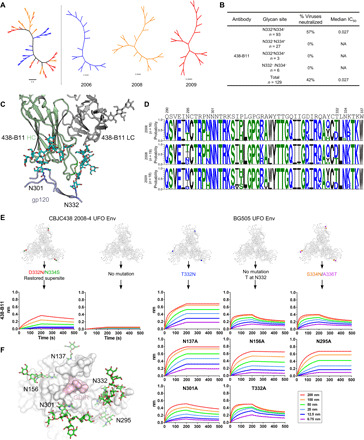
(A) NJ phylogenetic tree of 53 HIV-1 Env sequences isolated from donor CBJC438 by SGA. (B) Percent neutralization breadth and median IC50 in μg/ml were determined at an IC50 cutoff of 10 μg/ml for 438-B11. Viruses were separated into those containing an N-linked glycan at N332 but not N334 (N332+N334−), at N334 but not N332 (N332−N334+), at both N332 and N334 (N332+N334+), and at neither N332 nor N334 (N332−N334−). NA, not applicable. (C) Interaction of 438-B11 with the N332 and N301 glycans. (D) Sequence logo of the V3 region from three time points. (E) Binding affinity of 438-B11 antibodies to glycan mutants of CBJC438 UFO Env trimer and BG505 UFO Env trimer. Sensorgrams were obtained from an Octet RED96 instrument using a trimer titration series of six concentrations (200 to 6.25 nM by twofold dilution). For the CBJC438 Env, the D332N/N334S mutation restored the N332 supersite, whereas for the BG505 Env, the T332N mutation restored the N332 supersite. For BG505, a panel of glycan mutants was used to determine which glycan(s) surrounding the N332 supersite may be involved in epitope recognition. (F) Glycans surrounding the N332 supersite on the surface of BG505 Env trimer. A top view of the BG505 UFO.664 trimer surface with the GDIR motif colored pink. Glycans N137, N156, N295, N301, and N332 are represented with stick models and labeled.
