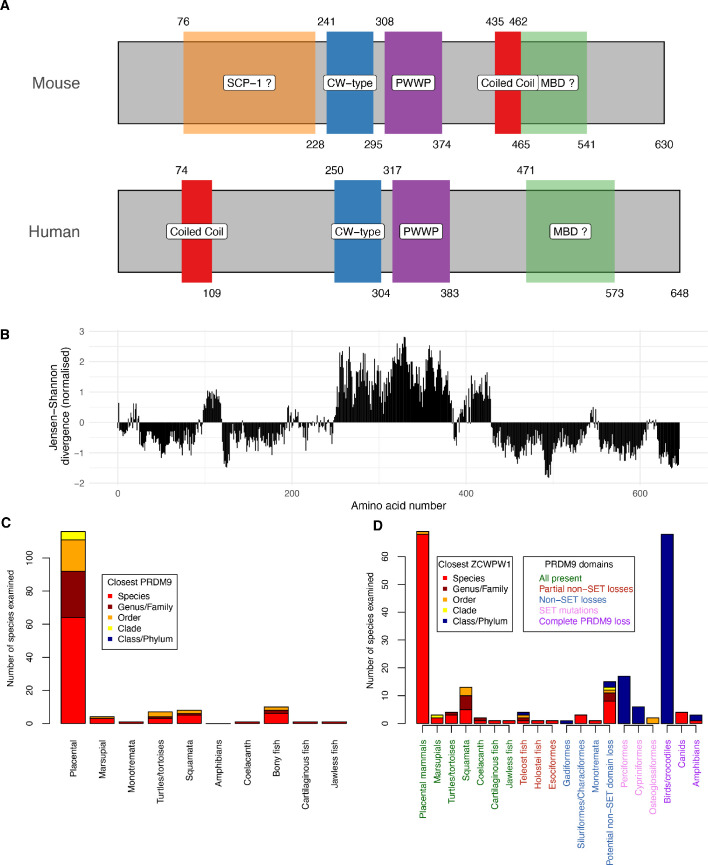
Figure 1. Domain organisation (A) and evolutionary conservation (B) of ZCWPW1, and co-evolution with PRDM9 (C,D).
(A) Protein domains in the human and mouse proteins (source: UniProt). Amino acid start and end positions of each domain are shown above and below the rectangles, respectively. Prediction of SCP-1 (SYCP1) domain from Marchler-Bauer and Bryant, 2004 and of MBDs (methyl-CpG binding domain) from Lobley et al., 2009 (Materials and methods). (B) Conservation of human amino acids, normalised Jensen-Shanon divergence normalised to mean of 0 and standard deviation of 1 is shown on the y-axis (a measure of sequence conservation, see Capra and Singh, 2007 and Johansson and Toh, 2010) computed from using multiple alignment of 167 orthologues (Materials and methods). (C) All species we identified as possessing ZCWPW1 copies were phylogenetically grouped into clades as previously (Baker et al., 2017) (x-axis) and each clade divided (stacked bars) according to whether ZCWPW1-possessing species within it also possess PRDM9 (‘Species’, red) or instead their closest PRDM9-possessing relative is respectively in the same genus/family, order, clade or order/phylum, with colours as given in the ‘Closest PRDM9’ legend. (D) As (C), but now showing the closest relative possessing ZCWPW1 (‘Closest ZCWPW1’ legend) for species possessing complete, partial or no identified PRDM9 copies. As in (C), the x-axis groups species into clades, now further divided based on data from Baker et al., 2017 into subclades according to the domains of PRDM9 lost or mutated across that subclade in all observed copies, reflecting multiple partial losses of particular PRDM9 domains, or complete loss of all PRDM9 copies (Main text). The x-axis labels are ordered and coloured according to the PRDM9 domains present (‘PRDM9 domains’ legend, where ‘SET’ refers to PRDM9’s PR/SET domain and the KRAB and SSXRD domains are grouped as ‘non-SET’, and ‘partial’ losses are seen in some but not all PRDM9 copies in that species). Further details are presented in Materials and methods, and the raw data in Figure 1—source datas 1 and 2.