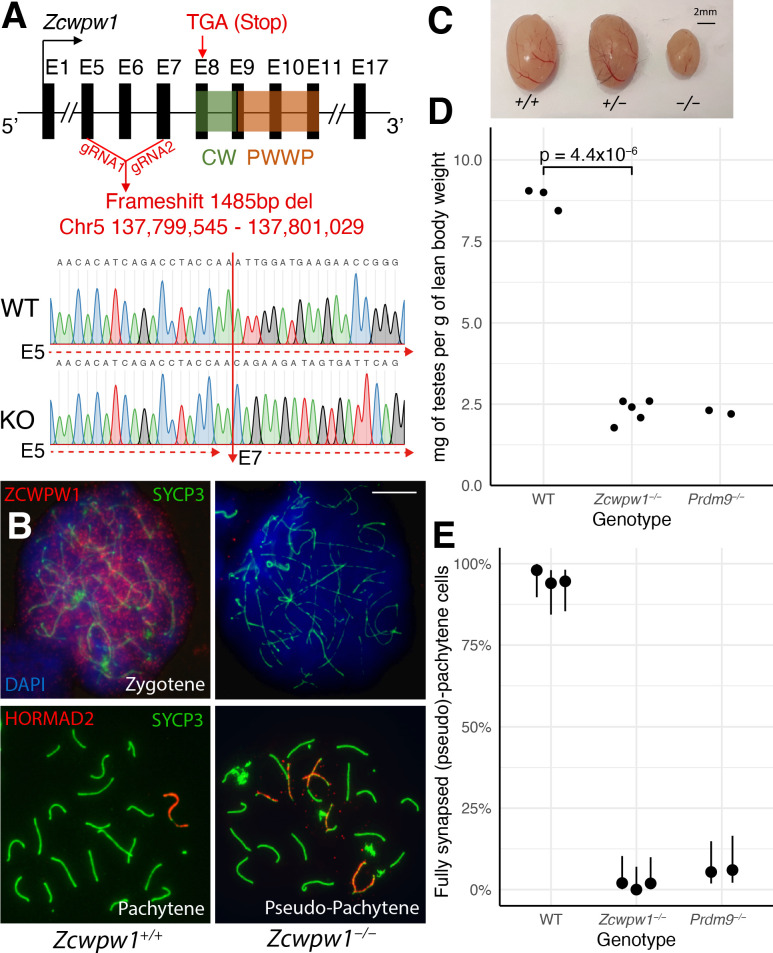
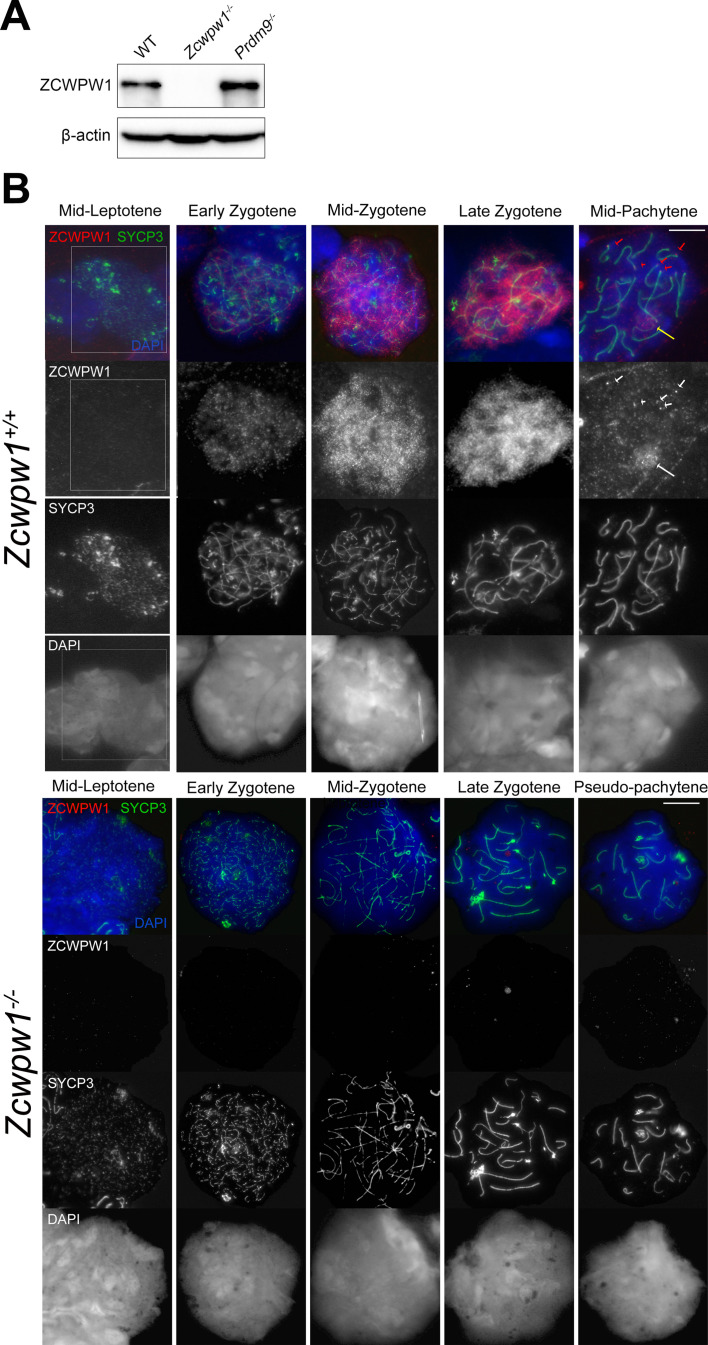
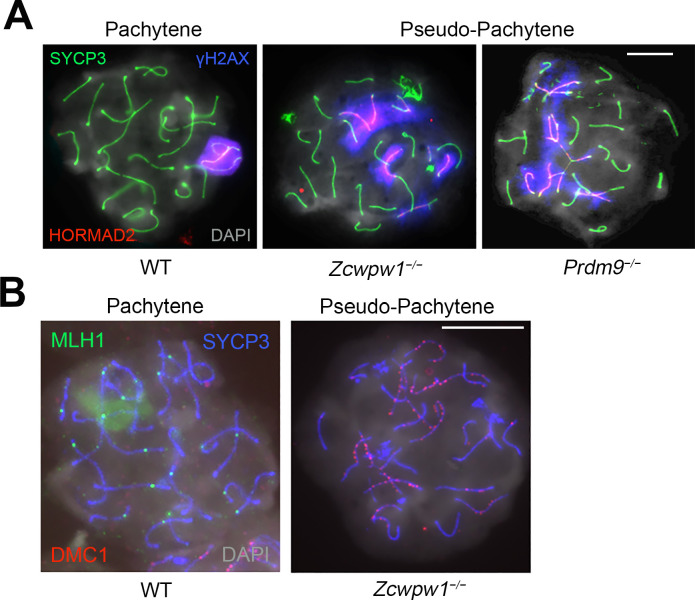
Figure 3. Zcwpw1−/ male mice show reduced testis size and asynapsis, similar to the Prdm9−/− mutant.
(A) Schematic of the Zcwpw1 knockout (KO) mouse line. E: Exon. gRNA: guideRNA. Sanger sequencing DNA chromatograms of wild-type (WT) and KO mice encompassing the deletion are shown. The intron-exon organisation is not to scale. (B) Immunofluorescence staining of testis nuclear spreads from 9- to 10-week-old Zcwpw1+/+ and Zcwpw1−/− mice for ZCWPW1 or HORMAD2 (red) which marks asynapsed chromosomes, and the synaptonemal complex protein SYCP3 (green) which labels the chromosome axis. Cells were counterstained with DAPI (blue) to visualise nuclei (top images). These images are representative of the data obtained for three mice per genotype. Scale bar: 10 μm. (C) Representative testes from 9- to 10-week-old WT (+/+), Het (+/−) and Hom (−/−) Zcwpw1 KO mice are shown. (D) Paired testes weight was normalised to lean body weight. Each datapoint represents one mouse. The p-value is from Welch’s two sided, two sample t-test. Raw data in Figure 3—source data 1. (E) Synapsis quantification in testis chromosome spreads immunostained with HORMAD2, as in (B). The percentage of mid-Pachytene (WT) or pseudo-Pachytene (Zcwpw1−/− and Prdm9−/−) cells with all autosomes fully synapsed is plotted by genotype; each datapoint represents one mouse, each with n≥ 49 cells analysed. Vertical lines are 95% Wilson binomial confidence intervals. Raw data in Figure 3—figure supplement 2—source data 1.