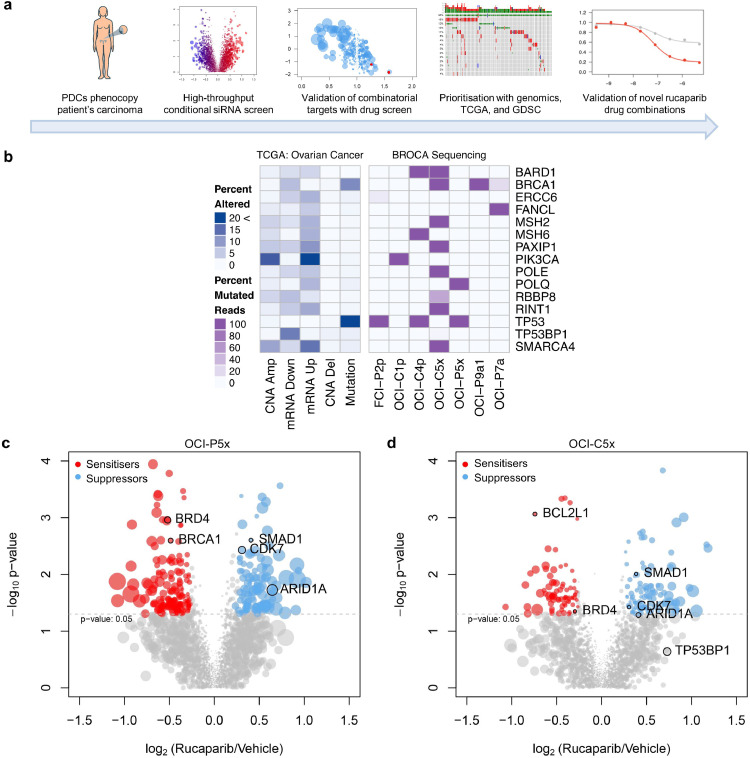
Fig. 1.
Integration of functional genomics with PDCs and big data for drug combination identification. (a) Schematic of the process used to identify rucaparib drug combinations, employing high-throughput conditional siRNA and drug screening on PDCs that genetically match the original tumour with prioritisation of drug targets using TCGA and GDSC. (b) Next generation sequencing of a targeted gene panel; only mutated genes within the PDCs are shown; darker purple indicates 100% mutated reads whereas lighter shades are a gradient of the percent mutated. (c) Volcano plots of the conditional siRNA screens on OCI-P5x cells. (d) Volcano plots of the conditional siRNA screens on OCI-C5x cells. Genes identified as rucaparib sensitisers are highlighted in red and genes identified as rucaparib suppressors in blue. Size of the dots represent the absolute difference in percent viability between siRNA alone and siRNA and rucaparib.