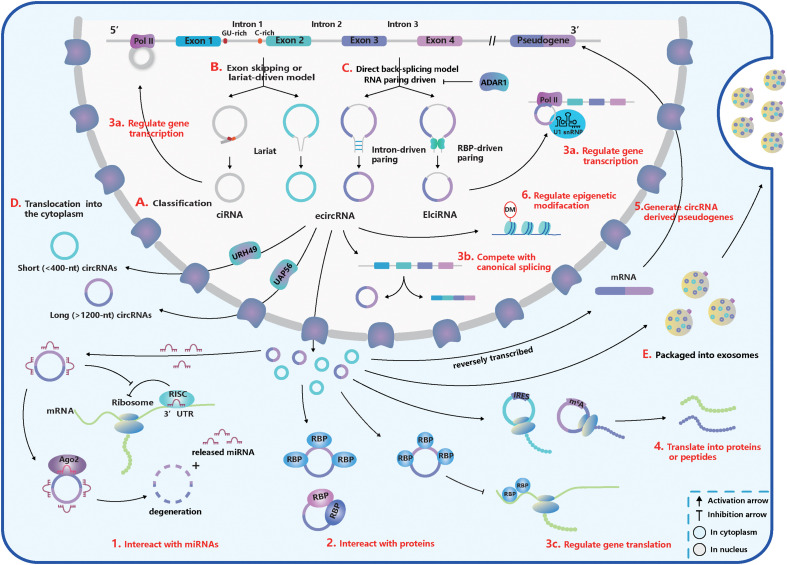
FIGURE 1.
The basic acknowledgment of circular RNA (circRNA). (A) CircRNA classification. CircRNAs can be classified into intronic circRNA (ciRNA), exonic circRNA (ecircRNA), and exon-intron circRNA (ElciRNA). (B) Exon skipping or lariat-driven mechanism of circRNA biogenesis. Lariats contained skipping exons and introns, can produce ecircRNAs and EIciRNAs. And ciRNAs are produced from a lariat intron with special GU-rich element at 5′ splice sites (ss) and C-rich element at 3′ branchpoint site. (C) Direct back-splicing or RNA pairing-driven mechanism of circRNA biogenesis. Special elements in introns like inverted repetitive Alu elements and reverse complementary matches (RCMs) and special RNA-binding proteins (RBPs) like NF90/110, quaking (QKI), and muscleblind (MBL) can facilitate the RNA-pairing. But adenosine deaminase acting on RNA 1 (ADAR1) can inhibit the RNA-pairing and reduce circRNA expression. (D) The translocation of circRNAs from the nucleus into the cytoplasm. Short (<400 nt) and long (>1,200 nt) circRNAs were transported into the cytoplasm based on the URH49 and UAP56, respectively. (E) Packaged into exosomes. CircRNAs can execute the regulatory function by exporting exosomes. (1–6) The function of circRNAs. (1) Interact with miRNAs. CircRNAs can work as miRNA sponges and promote the translation of miRNA-targeted genes. But some miRNAs can trigger the circRNA degeneration in the Ago2-slicer-dependent manner, which leads to the release of binding miRNAs. (2) Interact with proteins. CircRNAs can serve as transporters, decoys, or scaffolds for proteins and regulate their activities. (3a–c) Regulate gene transcription, splicing, and translation. (3a) CircRNAs can inhibit gene translation by competing for special RBPs. (3b) CiRNA and EcliRNA can promote parent gene transcription by interacting with Pol II. (3c) CircRNA biogenesis competes with canonical splicing and circRNAs like circMbl can regulate gene splicing. (4) Translate into proteins or peptides. CircRNAs like circZNF609 have internal ribosome entry site (IRES) within their sequences and can be translated. CircRNA translation also can be driven by N6-methyladenosine (m6A) modification in a cap-independent manner. (5) Generate pseudogenes. (6) Regulate epigenetic alterations. CircRNAs can adjust epigenetic modifications, such as DNA or histone methylation.