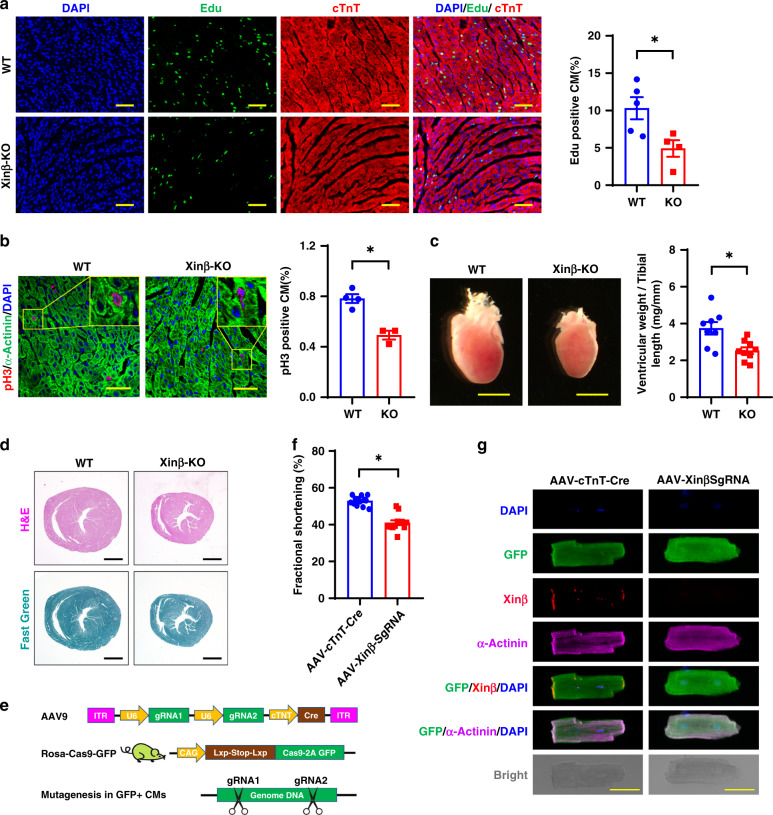
Fig. 2. Decreased cardiomyocyte proliferation in XinβKO hearts.
a Edu incorporation in P7.5 XinβKO and control hearts. DAPI marks nuclei, cTnT labels cardiomyocytes. Quantification in right panel. Scale bars = 100 µm. N = 4 and 5 biologically independent samples, respectively, *P < 0.05. b pH3 staining of P7.5 XinβKO and control hearts. DAPI marks nuclei, α-actinin labels cardiomyocytes. Quantification in right panel. Scale bars = 40 µm. N = 4 and 3 biologically independent samples, respectively, *P < 0.05. c Heart morphology (left panel) and ventricle weight vs. tibial length ratio (right panel) of P7.5 XinβKO and control hearts. Scale bars = 2 mm. N = 9 and 8 biologically independent samples, respectively,*P < 0.05. d Hematoxylin and eosin (red) and Fast Green (green) staining of p7.5 XinβKO and control hearts. Scale bars = 1 mm. e AAV9-based CRISPR/Cas9 strategy to mutate the Xinβ gene in postnatal mouse hearts; f percent fractional shortening (FS) change in 4-week-old AAV-cTnT-Cre and AAV-Xinβ-sgRNA-treated mice. N = 13 and 12 biologically independent samples, respectively, *P < 0.05. g Morphology and immunostaining of isolated cardiomyocytes from hearts of adult XinβKO and control mice. Scale bars = 50 µm.