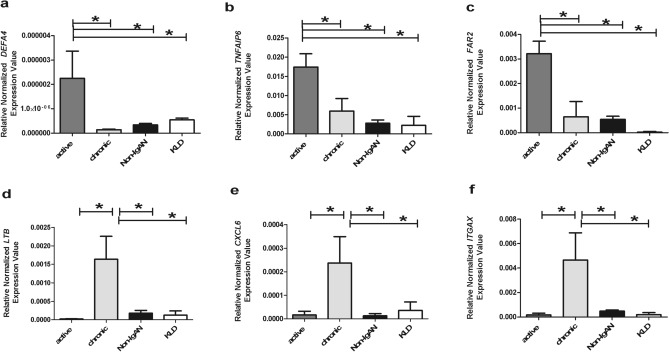
Figure 3.
Gene expression levels evaluated by real-time (RT)PCR in kidney biopsy specimens of IgAN patients with active (DEFA4, TNFAIP6, FAR2) and chronic renal lesions (LTB, CXCL6 and ITGAX), 22 non-IgAN (non-IgAN) and 7 kidney living donors (KLD). (A–C) DEFA4, TNFAIP6, FAR2 normalized gene expression levels are significantly higher in IgAN patients with active renal lesions compared to all other groups. (A) DEFA4 ANOVA F(3,19) = 8.012, p = 0.0012; Tukey’s multiple comparison test, active versus chronic, active versus non IgAN and active versus KLD all p < 0.05. (B) TNFAIP6 ANOVA F(3,21) = 6.761, p = 0.0023; Tukey’s multiple comparison test, active versus chronic, active versus non IgAN and active versus KLD all p < 0.05. (C) FAR2 Kruskal–Wallis test p = 0.0013. Dunn’s multiple comparison test, active versus chronic, active versus non-IgAN and active versus KLD, all p < 0.05. (D–F) LTB, CXCL6, ITGAX normalized gene expression levels are significantly higher in IgAN patients with chronic renal lesions compared to all other groups. (D) LTB ANOVA F(3,16) = 5.646 p = 0.0079. Tukey’s multiple comparison test, chronic versus active, chronic versus non IgAN , chronic versus KLD all p < 0.05. (E) CXCL6 Kruskal–Wallis test p = 0.0057. Dunn’s multiple comparison test, chronic versus active, chronic versus non-IgAN and chronic versus KLD, all p < 0.05. (F) ITGAX Kruskal–Wallis test p = 0.0009. Dunn’s multiple comparison test, chronic versus active, chronic versus non-IgAN and chronic versus KLD, all p < 0.05.