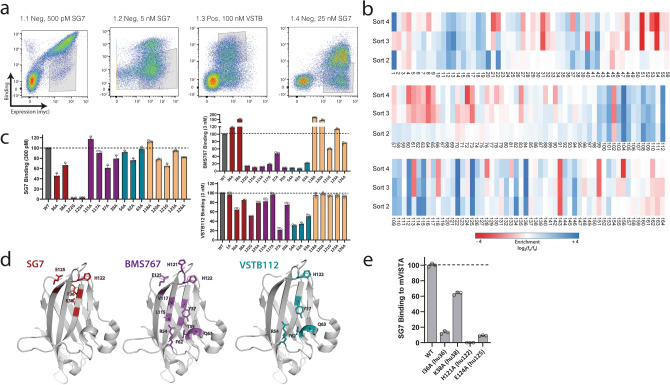
Figure 3.
SG7 binds to a distinct epitope on VISTA. (a) Library screening progression used to isolate human VISTA mutants that lost binding to SG7 but retained binding to VSTB112. Flow cytometry gates used in screening are shown on dot plots of individual sorts. (b) Enrichment of mutations at each residue location in human VISTA obtained from NGS analysis of gated populations after each epitope mapping sort (blue—more enriched, red—less enriched). (c) Analysis of individual human VISTA mutations expressed on yeast. Binding intensity of each mutant to SG7, BMS767, or VSTB112 at the respective approximate dissociation constant (Kd) of each antibody. Bars are colored based on predicted epitope (SG7-red, BMS767-purple, VSTB112-cyan, neighboring residues-beige). Binding signal of each mutant was normalized to wild-type hVISTA. (d) Epitopes of SG7 (red), BMS767 (purple), and VSTB112 (cyan) antibodies based on single clone binding analysis. (e) Analysis of individual mouse VISTA mutations displayed on yeast. Selected mutants correspond with aligned human VISTA residues (in parentheses) that make up or are near the predicted SG7 epitope. Binding intensity of each mutant to 4 nM SG7 is shown. Binding signal of each mutant was normalized to wild-type mVISTA. Mean ± SD for triplicate measurements are shown for (c,e).