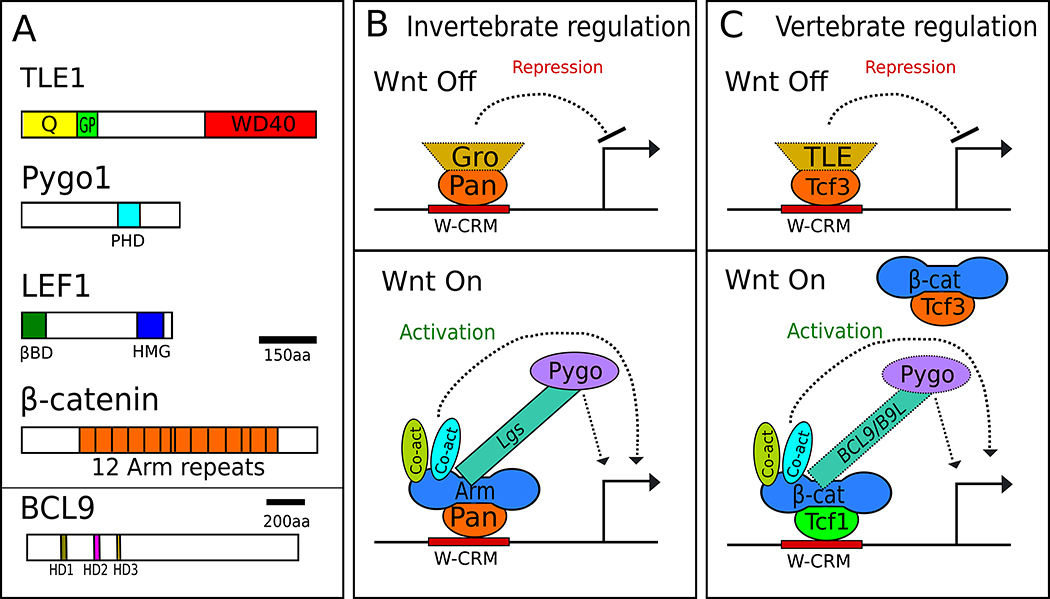
Fig 1. Traditional models of the Wnt transcriptional switch.
A) Domain architecture of the principal components of the switch, using human proteins of each family as archetypes. Colored portions represent interaction domains: Q – glutamine-rich domain, GP – Glycine/Proline-rich domain, PHD – PHD zinc finger domain, βBD – β-catenin binding domain, HMG – High Mobility Group, PBD – Pygopus binding domain, HD1-3 – Homology Domains 1-3 B) The transcriptional switch model in invertebrates. In the absence of Wg signaling in Drosophila, Pan recruits Gro to W-CRMs and represses transcription. When Wg signaling is activated, Gro is displaced and Arm binds to Pan, recruiting Lgs, Pygo and other co-activators, which activate transcription. Lgs acts as an adaptor between Arm and Pygo. C) The TCF switch model in vertebrate Wnt signaling. In the absence of Wnt signaling, repressive TCFs such as TCF3 are bound to W-CRMs. TLEs likely plays a role for repression of many targets. Upon Wnt stimulation, TCF3 is replaced by activating TCFs such as TCF1, which recruits β-catenin and other co-activators. Some factors in panels B and C are depicted with dotted lines to highlight the likelihood that they likely do not regulate all Wnt targets in their respective systems.