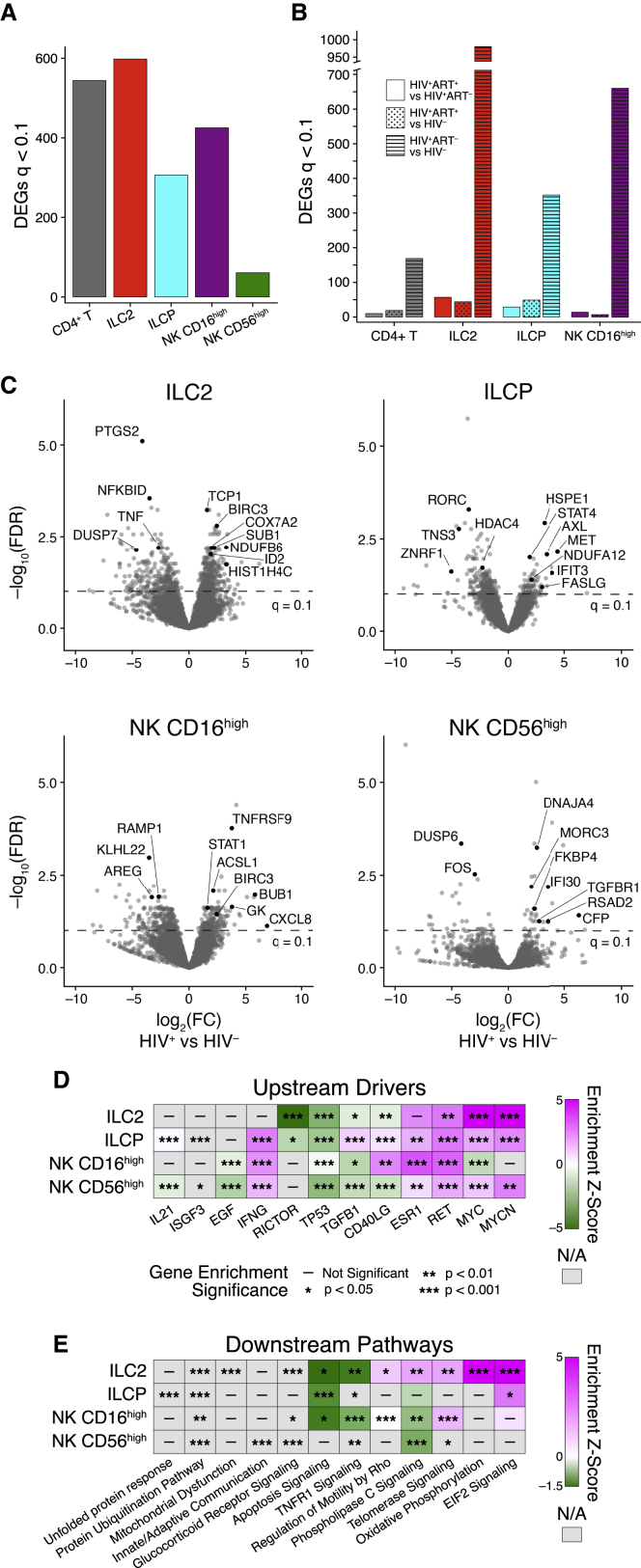
Figure 4.
Blood ILC Subsets Are Transcriptionally Activated during Chronic Pediatric HIV
(A and B) Number of DEGs in whole transcriptomes of CD4+ T cells, ILC2s, ILCPs, CD56high, and CD16high NK cells between HIV-infected and HIV-uninfected pediatric subjects (A) and among HIV-uninfected, HIV-infected treated and HIV-infected untreated pediatric subjects (B) (see Table S1 for subject numbers). DEGs were called using DESeq2 with a significance cut-off of FDR < 0.1.
(C) Volcano plots of the DEGs between HIV-infected (positive) and HIV-uninfected (negative) pediatric subjects in (A). Genes of interest are annotated with black dots; see Table S3 for all DEGs. Dotted line annotates the significance cut-off of FDR q < 0.1.
(D and E) Select upstream drivers (D) and canonical pathways (E) significant in ingenuity pathway analysis (IPA) of DEGs from each ILC subset. For directionally annotated pathways, a Z-score is calculated to represent up- or downregulation of the driver or pathway. If a driver or pathway is not directionally annotated in IPA, or there are not enough genes in the list to calculate a Z-score, N/A is reported. See Table S4 for the full IPA results. See also Figure S2.