Figure 6.
Coordinated Transcriptional ILC3 and NK Cell Response in the Pediatric HIV-Infected Tonsil
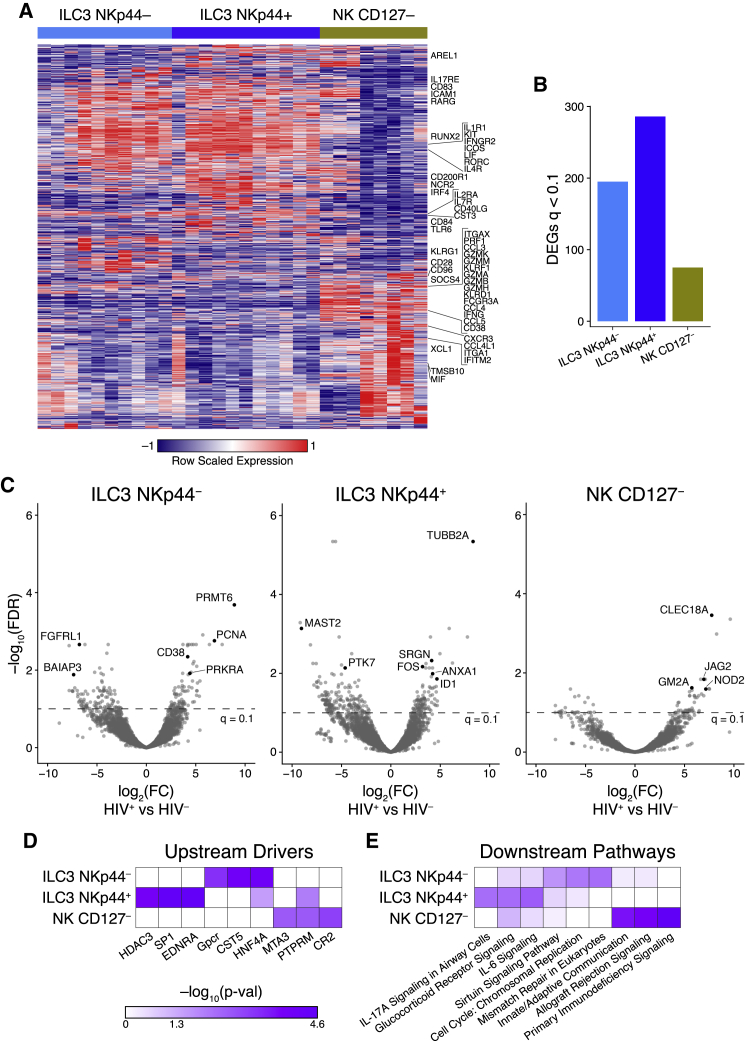
(A) Heatmap showing DEGs among ILC3 NKp44–, ILC3 NKp44+, and NK CD127– subsets from four HIV-negative pediatric tonsils performed in duplicate or triplicates with canonical genes annotated.
(B) Number of DEGs in whole transcriptomes of tonsils of ILC3 NKp44–, ILC3 NKp44+, and NK CD127– cells among viral-suppressed (<20 HIV RNA copies/ml plasma), HIV-infected (HIV+; n = 3), and gender- and age-matched HIV-uninfected pediatric subjects (HIV–; n = 4).
(C) Volcano plot showing significance plotted against log2 fold change for DEGs in the ILC3 NKp44–, ILC3 NKp44+, and NK CD127– populations with genes upregulated in HIV-infected tonsils shown as positive log2 fold change (right) and genes downregulated by HIV infection shown as negative log2 fold change (left). Genes of interest are annotated with black points and with dotted line annotating the significance cut-off of FDR q < 0.1.
(D and E) Upstream molecules predicted to be involved in initiating pathways shown in (E) determined by ingenuity pathway analysis of DEGs from the each of the three innate tonsil lymphocyte subsets. See also Figure S4.