Figure 6.
SUMOylation of Dppa2 and Dppa4 Inhibits the Transition to 2C-like Cells and Transcript Activation
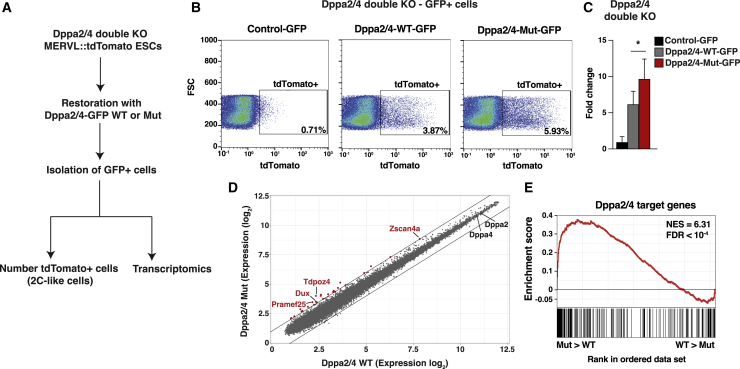
(A) Experimental procedure used for counting the number of 2C-like cells and for transcriptomic analysis.
(B) Flow cytometry profiles showing expression of the Mervl::tdTomato reporter (x axis) in Dppa2−/− and Dppa4−/− double-knockout (KO) ESCs transfected with GFP (left), Dppa2-WT-GFP + Dppa4-WT-GFP (middle), or Dppa2-Mut-GFP + Dppa4-Mut-GFP (right). The population and percentage of tdTomato-positive cells are shown in the square. Representative example, n = 3.
(C) Percentage of tdTomato-positive cells in Dppa2−/− and Dppa4−/− double-KO ESCs complemented with GFP, Dppa2-WT-GFP + Dppa4-WT-GFP, or Dppa2-Mut-GFP + Dppa4-Mut-GFP. Error bars indicate mean + SD, n = 3.
(D) Scatterplot comparing gene expression of double-KO ESCs complemented with Dppa2-WT-GFP + Dppa4-WT-GFP or Dppa2-Mut-GFP + Dppa4-Mut-GFP. Cells were sorted for GFP expression in both conditions. Red dots indicate overexpressed transcripts (fold change > 2). n = 3.
(E) Gene set enrichment analysis (GSEA) for Dppa2/4-specific target genes in double-KO ESCs complemented with Dppa2-WT-GFP + Dppa4-WT-GFP or Dppa2-Mut-GFP + Dppa4-Mut-GFP. For x axis, genes were ranked on the basis of the ratio of Dppa2/4-Mut-GFP versus Dppa2/4-WT-GFP. NES, normalized enrichment score; FDR, false discovery rate.