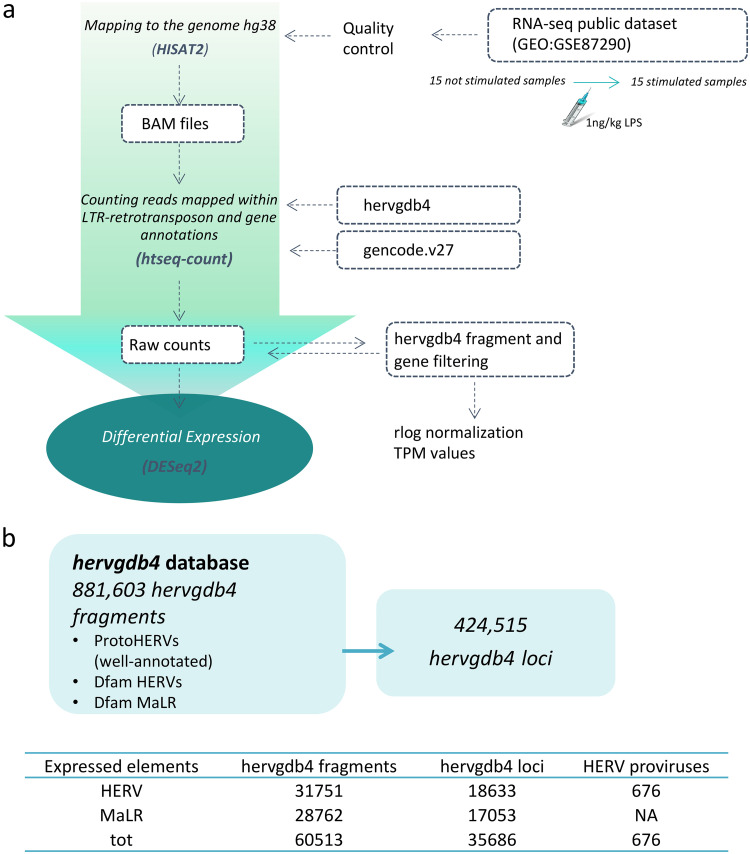
FIG 1.
Experimental design of differential expression analysis. (a) RNA-Seq workflow for the identification of modulated HERVs and MalRs. The input files used are given in blue boxes. (b) Scheme showing the composition of the hervgdb4 database. The amounts of expressed hervgdb4 fragments and loci were obtained by filtering the raw counts and are summarized in the table at the bottom of the figure. The coordinates of the expressed ReTe most-intact proviruses were obtained by using the findOverlaps function from package Iranges and the coordinates of the expressed hervgdb4 fragments.