Fig. 7.
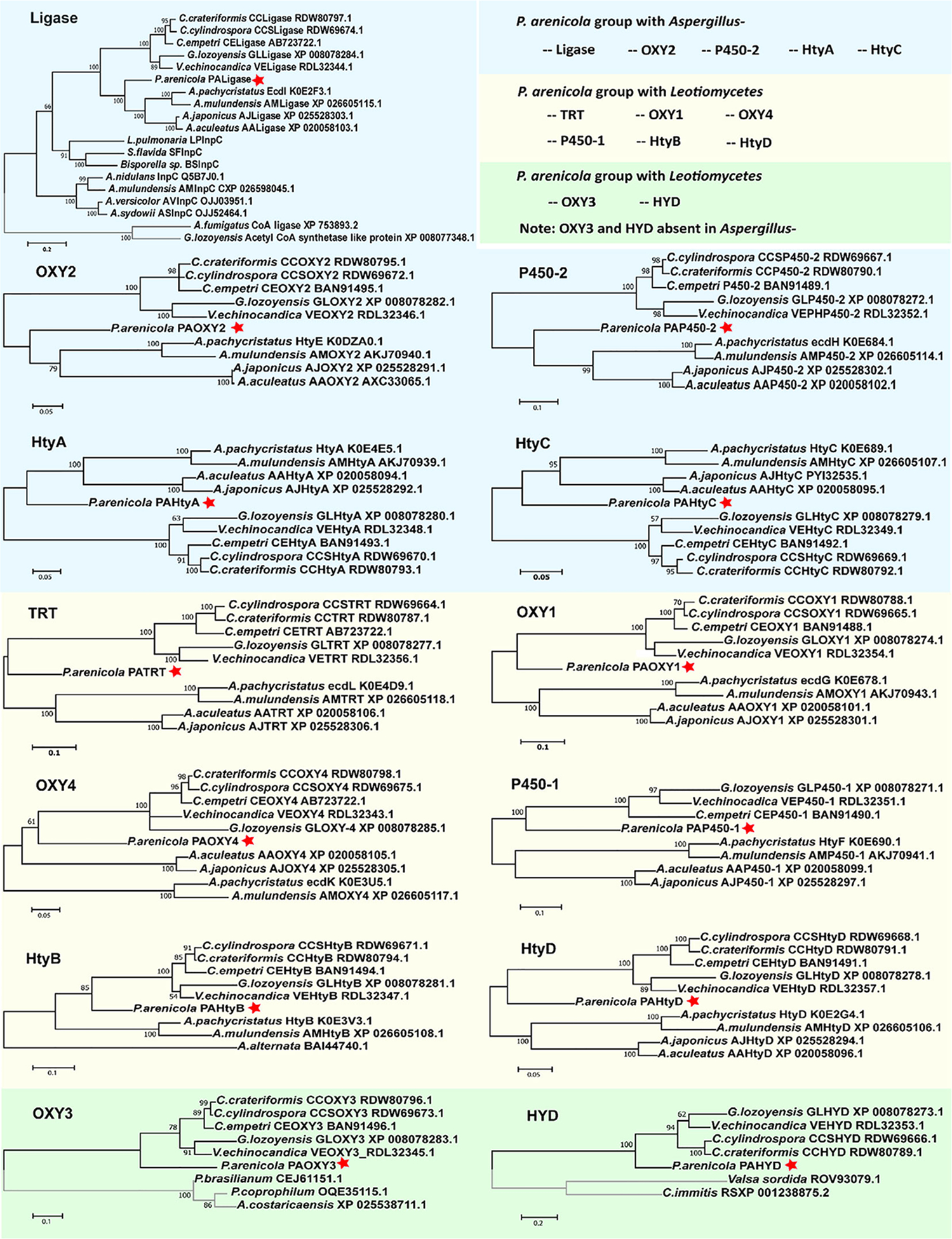
Abbreviated trees for maximum likelihood phylogenies inferred from genes in the echinocandin biosynthetic gene clusters. Maximum likelihood phylogeny was inferred from each of the echinocandin biosynthetic-related genes. Each of the echinocandin biosynthetic-related genes forms a monophyletic lineage. Eight of 13 P. arenicola acrophiarin biosynthetic genes group with genes Leotiomycete-type echinocandins. A. Phylogeny of acyl-AMP and AMP-dependent ligases. B, G, H and L. Phylogeny of non-heme iron, α-ketoglutarate-dependent oxygenases. C and I. Phylogeny of cytochrome P450s. D. Phylogeny of 2-isopropylmalate synthases. E. Phylogeny of isopropylmalate dehydrogenases. F. Phylogeny of ABC transporters. J. Phylogeny of d-amino acid aminotransferases. K. Phylogeny of isopropylmalate dehydratases. M. Phylogeny of thioesterases. Numbers at nodes are likelihood support values.
