Fig. 8.
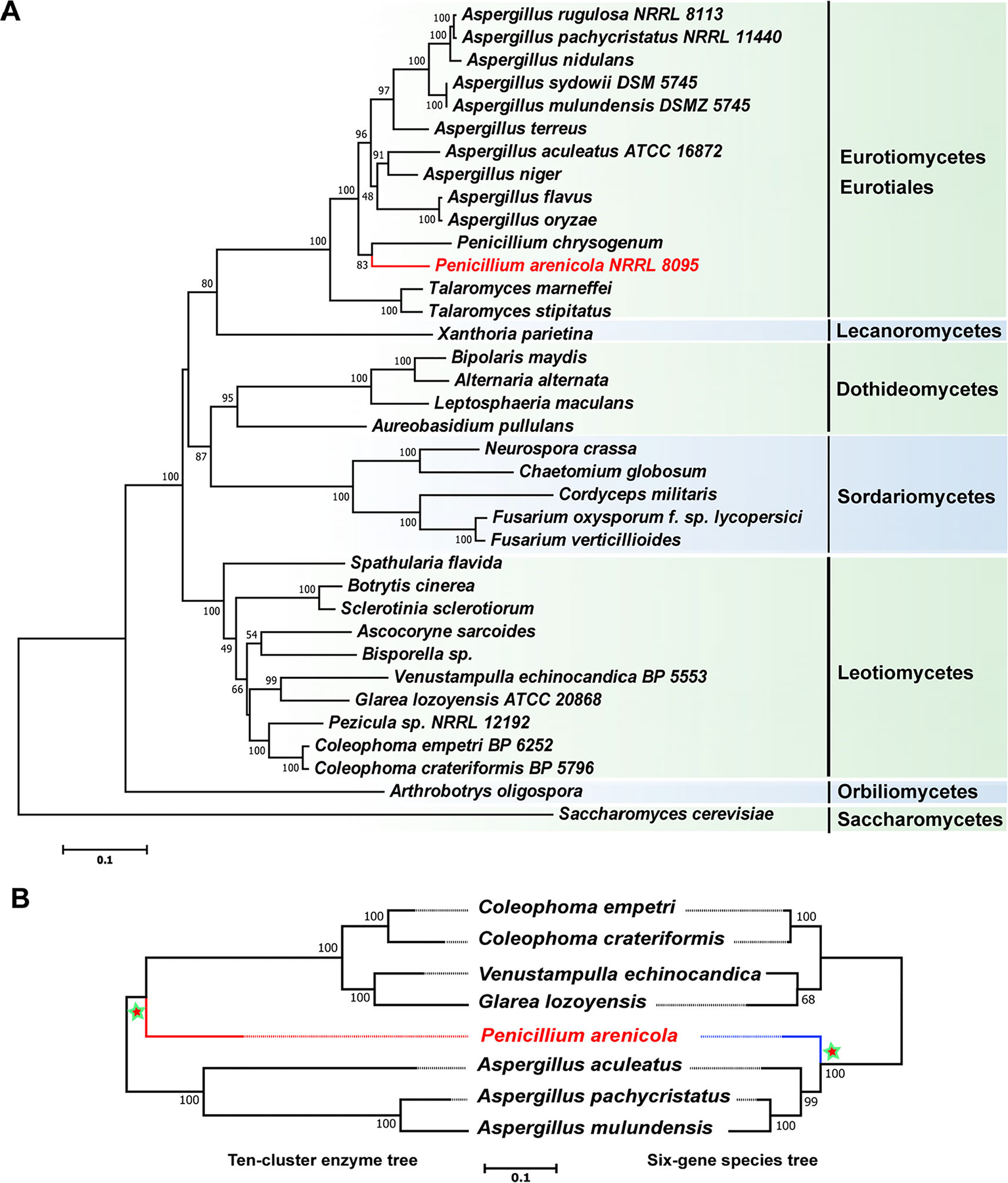
Maximum likelihood phylogenies of fungal species and the enzymes of the echinocandin pathway. A. Phylogenetic reconstruction of the echinocandin-producing fungi and other Ascomycetes using maximum likelihood analysis of a six-gene dataset consisting of DNA fragments of the 18S rRNA gene, the 28S rRNA gene, the ITS RNA region, the β-tubulin gene, the translation elongation factor 1-α gene and the RNA polymerase II subunit 2 gene. B. Consensus phylogenetic pattern for gene tree for 10 shared enzymes of all echinocandin pathways (left) and the established phylogenetic tree extracted from the data in panel A. Trees are rooted at the midpoint. Numbers at nodes are likelihood support values. The 10-cluster enzyme tree significantly conflicted with the species phylogeny tree (Shimodaira–Hasegawa test, P = 0; weighted Shimodaira–Hasegawa test, P = 0). Dotted lines are aids to connect branch tips to strain name.
