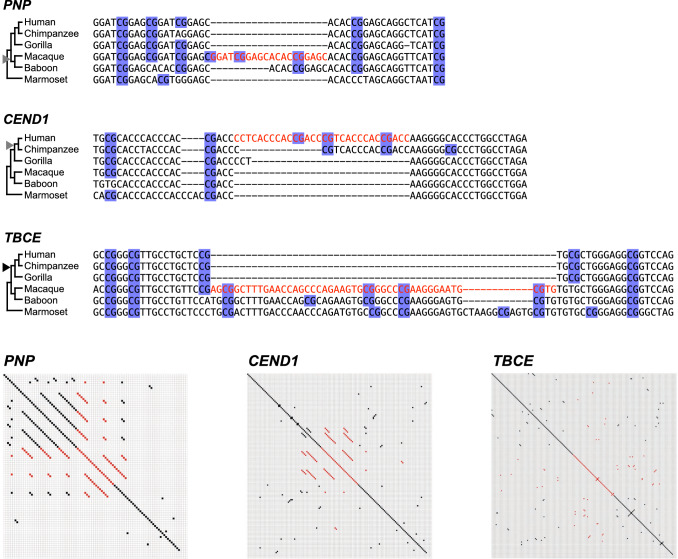
Fig. 5.
Examples of indels in human and macaque promoters. Sequence alignments are shown for the PNP, CEND1, and TBCE promoters, in which an indel altered the promoter types between the two species. Harr plots were drawn to detect tandem repeats related to insertion or deletion events (rheMac8 chr7:81,686,561–81,686,600 for PNP, hg19 chr11:790,324–790,373 for CEND1, and rheMac8 chr1:211,651,491–211,651,557 for TBCE). Each dot indicates a 5-base perfect match. Indel-related sequences are shown in red. Multiple alignments of the six primate species were drawn to determine the type of mutation event, namely insertion or deletion. The gray and black triangles indicate the occurrence of an insertion and deletion, respectively. CpG islands were gained in macaque PNP and human CEND1 through insertion and lost in hominoid TBCE via deletions. All CpG sites are shaded in blue