Fig. 10.
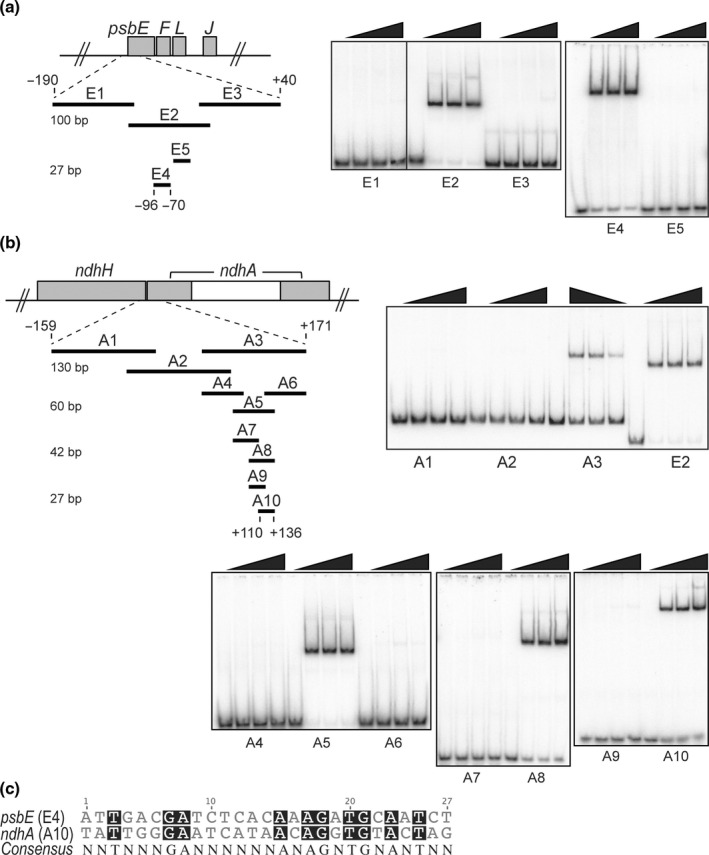
Preferential DNA binding of rMDA1 to regions near the 5′‐end of chloroplast psbE and ndhA Arabidopsis genes. (a) Gel mobility shift assays showing specific DNA binding of rMDA1 to a short DNA sequence upstream of psbE start codon. Different amounts of rMDA1 (0, 12.5, 25, 50 nM) were incubated with overlapping DNA segments of various sizes that span the −190/+40 genomic region from psbE start codon in the presence of poly(dI‐dC) competitor. The length of the DNA fragments (bp) is given on the left side. DNA fragment E2 was used for MDA1 DNase footprinting analysis shown in Supporting Information Fig S6. (b) Gel mobility shift assays showing specific DNA binding of rMDA1 to a DNA sequence downstream of ndhA start codon. Binding conditions were identical to those used in (a) and the DNA fragments covered a −159/+171 genomic region from the ndhA start codon. (c) Sequence alignment of the 27‐bp psbE and ndhA MDA1 binding sites. Conserved bases are shaded in black and the consensus sequence is given below.
