Fig. 11.
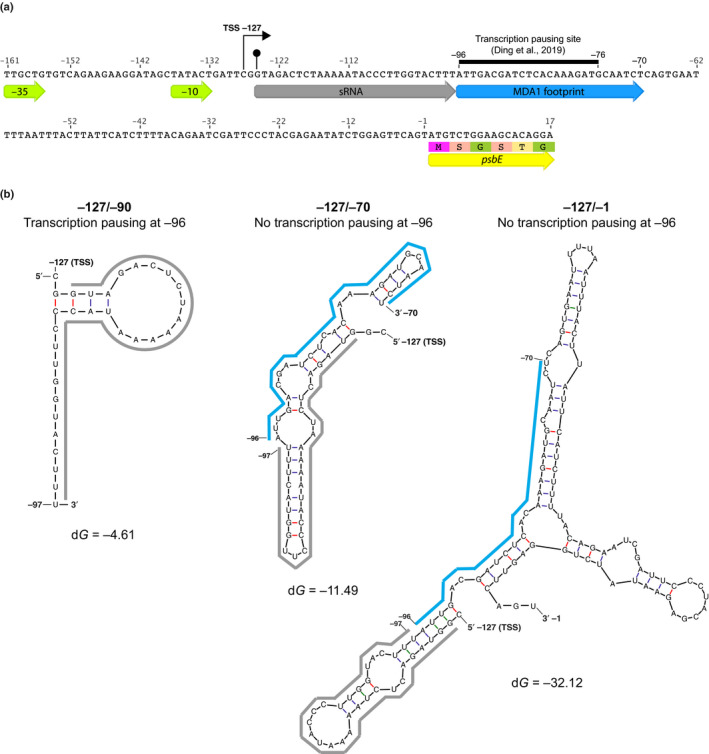
Potential mechanism of MDA1 action on psbE 5′ RNA stabilization in Arabidopsis. (a) Nucleotide sequence of the −161/+17 psbE genomic region. The genomic positions are given according to psbE start codon. The −35 and −10 consensus eubacterial plastid‐encoded RNA polymerase (PEP) promoter elements, transcription initiation site (TSS) (right squared arrow) and in vivo 5′‐end of processed psbE mRNA (circled arrow tip) are positioned on the map. The sequences of the 5′ psbE sRNA PPR footprint that is specifically lost in mda1 and MDA1 DNA binding site are underlined by grey and blue arrows, respectively. The MDA1 binding site contains a transcription pausing site (Ding et al., 2019). The 5′ region of the psbE coding sequence is underlined by a yellow arrow. (b) Mfold prediction of the most stable structure of neotranscribed RNA sequences from psbE TSS (−127) to the PEP pausing site (−96) or farther downstream to the MDA1 binding site (−70) or upstream the psbE start codon (−1). The sequences of the 5′ psbE sRNA and MDA1 footprint are underlined using the same color code as in (a). The calculated dG for each RNA structure is given in kcal mol–1. MDA1 DNA binding downstream of the PPR footprint sequence pauses gene transcription and would prevent the folding of the footprint into a secondary RNA structure that is deleterious for the PPR binding to the 5′ end of psbE mRNA and therefore, the post‐transcriptional stabilization of the processed psbE mRNA in vivo.
