Figure 1.
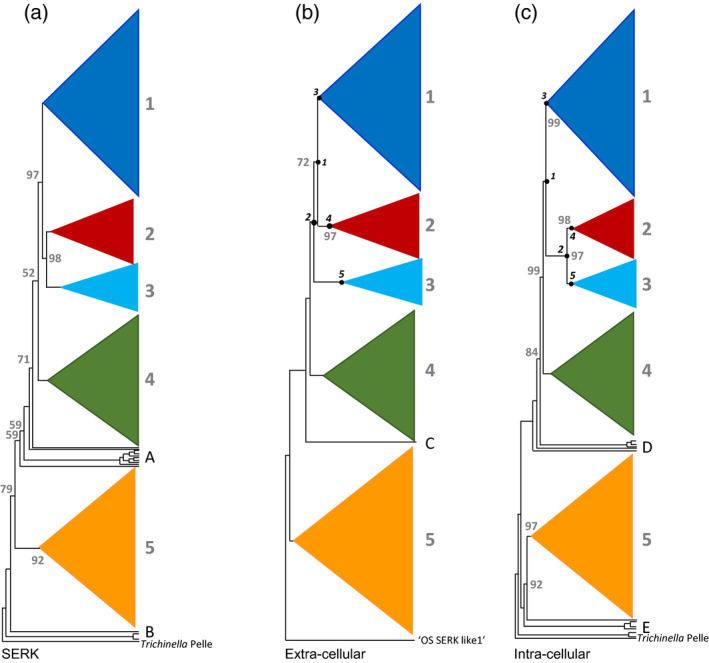
Maximum likelihood phylogenetic trees (cladogram style) of 1342 land plant leucine‐rich repeat receptor‐like kinase II (LRR‐RLKII) amino acid sequences, with five main clades (collapsed), labelled here as 1–5, and using the nematode Trichinella Pelle kinase sequence as the outgroup.
Trees are based on (a) the combined intracellular (‘Int’) + extracellular (‘Ext’) sequence matrix, (b) the extracellular LRR domain (matrix ‘Ext’, see text) rooted by inference from (a), and (c) the intracellular kinase domain (matrix ‘Int’). Bootstrap support values of less than 100 are indicated at the main nodes. Numbers in circles indicate nodes used in apomorphy reconstruction (see text). Terminals not belonging to any of the five main clades are: (A) Marchantia polymorpha [receptor kinase‐like SERK (RKS)2, RKS13, RKS0], Closterium ehrenbergii somatic embryogenesis receptor kinase (SERK), Physcomitrella patens (RKS8, RKS0), Selaginella moellendorffii, Adiantum capillus‐veneris; (B) Oryza sativa (SERK‐like 1); (C) M. polymorpha (RKS2); (D) P. patens (RKS8, RKS0), S. moellendorffii, A. capillus‐veneris; (E) Closterium ehrenbergii SERK, S. moellendorffii, M. polymorpha (RKS13, RKS0).
