Figure 5.
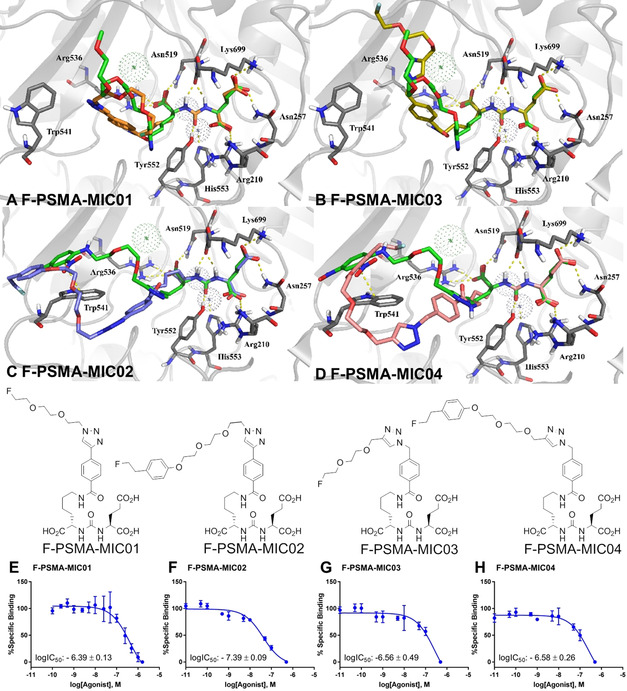
Molecular docking studies and binding affinities of the 2 nd generation F‐PSMA‐MIC compounds. A–D: Molecular docking poses. (A) F‐PSMA‐MIC01 (orange) and (B) F‐PSMA‐MIC03 (yellow), superimposed on the binding mode of MeO‐P4 with PSMA (PDB ID: 2XEJ); (C) F‐PSMA‐MIC02 (purple) and (D) F‐PSMA‐MIC04 (pink), superimposed on the binding mode of ARM‐P2 with PSMA (PDB ID: 2XEI). Protein is represented as grey cartoon with key residues in sticks, co‐crystallized ligands in green, metal ions as dotted spheres. Hydrogen bonds and π−π stackings are depicted as yellow dashed lines. (E–H) LogIC50 determination. Mean values ± SD (E,F and H: n=3, G: n=4). Competitive binding radioassays of the F‐PSMA‐MIC compounds on LNCaP cells using [18F]PSMA‐1007 as radioactive competitor.
