Figure 1.
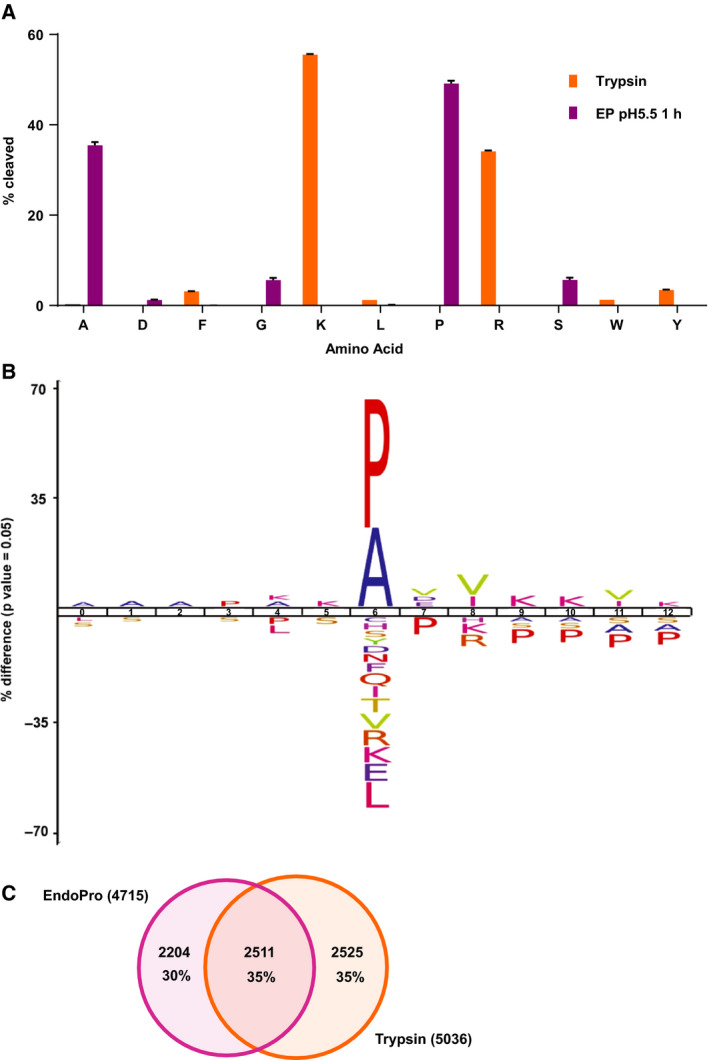
Characterization of EndoPro cleavage specificity. (A) Overview of amino acids after which was cleaved by EndoPro (n = 4, purple) and trypsin (n = 4, orange), based on a nonspecific search, revealing a high specificity of 84.6% A/P and 89.6% R/K for EndoPro and trypsin, respectively. Only amino acids with a cleavage frequency of 1% or higher were included. Data are represented as mean percentage of total cleavages per protease ± SEM. (B) An iceLogo showing the differences between the EndoPro cleavage site environment (17 032 unique environments from nonspecific search) and the human proteome, illustrating a disfavor for R/K on the +2 position and a reluctance to cleave between proline residues. (C) Overlap of unique proteins identified by EndoPro or trypsin using a semispecific search. Although the sizes of the identified proteomes are roughly equal, the overlap between the two is only 35%.
