Figure 7.
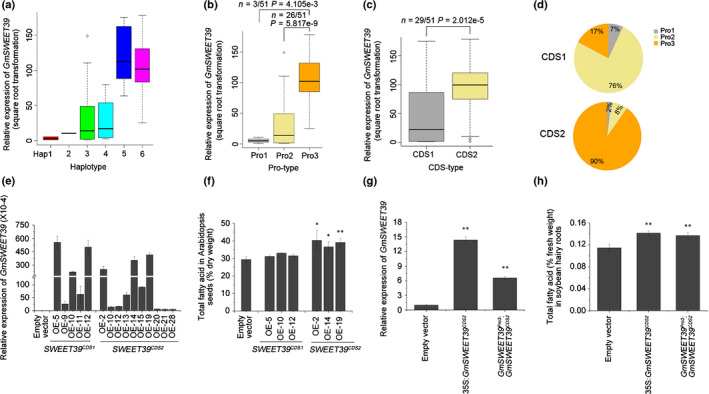
Allelic effects of GmSWEET39 in the representative cultivated soybean (Glycine max) population, transgenic Arabidopsis and soybean hairy roots. (a–c) Comparison of GmSWEET39 expression levels between soybean accessions containing different haplotypes (Hap), promoters (Pro) and coding sequences (CDS) of GmSWEET39, respectively. The dataset is the same as that in Fig. 5(c). The central bold line within the box represents the median; box edges indicate the upper and lower quantiles; whiskers show the 1.5 × interquartile range and points indicate outliers. P‐values were determined by two‐tailed two‐sample Wilcoxon tests. (d) Frequencies of different GmSWEET39 Pro‐types in two CDS groups among 80 cultivated soybean accessions. (e) Expression levels of GmSWEET39 CDS1 and GmSWEET39 CDS2 in transgenic Arabidopsis. AtACTIN7 was used as the internal control and the relative expression of GmSWEET39 was calculated by 2‐∆Ct (∆Ct = CtGmSWEET39 − CtAtACT7). (f) Total fatty acid (TFA) content in Arabidopsis seeds. Three independent transgenic lines overexpressing (OE) GmSWEET39 CDS1 or GmSWEET39 CDS2 were analyzed. (g) Relative expression of GmSWEET39 in transgenic soybean hairy roots. CaMV 35S promoter or SWEET39 Pro3 were used to express GmSWEET39. GmUKN1 was used as the internal control and the relative expression of GmSWEET39 was normalized to that in soybean hairy roots transformed by the empty vector. (h) The TFA content in soybean hairy roots. Data represent mean ± SD (n ≥ 3). Significant differences between GmSWEET39 transgenic lines and empty vector transgenic lines: *, P = 0.05; **, P = 0.01 (Student’s t‐test). SWEET, Sugars Will Eventually be Exported Transporter.
