Figure 2.
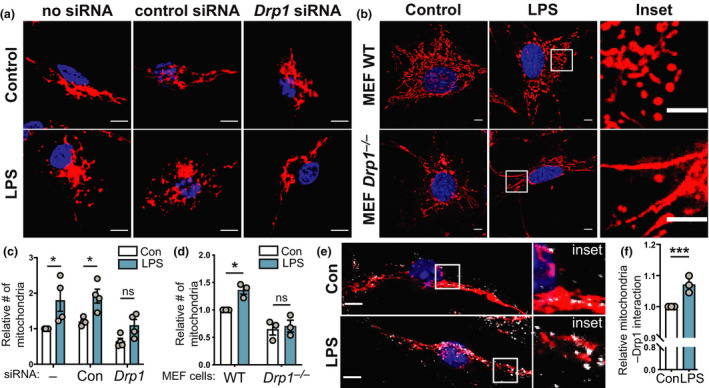
LPS‐inducible mitochondrial fragmentation requires Drp1. (a–f) The indicated cell populations were stimulated with LPS (100 ng mL−1) or left untreated for 6 h, prior to performing indicated analyses. Mitochondrial numbers were determined relative to the unstimulated control in each experiment. (a) BMMs were electroporated with no siRNA or siRNAs targeting either a control gene (Hdac1) or Drp1, incubated for 18 h and then treated with LPS. Cells were stained with MitoTracker (mitochondria, red) and DAPI (nuclear DNA, blue). (b) WT and Drp1 −/− MEF cells were treated with LPS, before being stained with MitoTracker (mitochondria, red) and DAPI (nuclear DNA, blue). Images in a and b are representative of a minimum of three independent experiments. (c, d) Quantification of the relative number of mitochondria in a and b. (c) Mean ± s.e.m. of four independent experiments (calculated using two‐way ANOVA with Šidák’s multiple comparisons test) and (d) mean ± s.e.m. of three independent experiments (calculated using two‐way ANOVA with Šidák’s multiple comparisons test). (e) BMMs were LPS treated before being stained with MitoTracker (mitochondria, red), DAPI (nuclear DNA, blue) and anti‐Drp1 (white). Images are representative of three independent experiments. (f) Relative colocalization of Drp1 with MitoTracker, with data displayed relative to unstimulated control cells (mean ± s.e.m., three independent experiments, one sample t‐test). In all panels, *P < 0.05, ***P < 0.001. Scale bars, 5 µm. BMMs, bone marrow‐derived macrophages; Con, control; DAPI, 4′,6‐diamidino‐2‐phenylindole; LPS, lipopolysaccharide; MEF, mouse embryonic fibroblast; ns, not significant; siRNA, small interfering RNA; WT, wild type.
