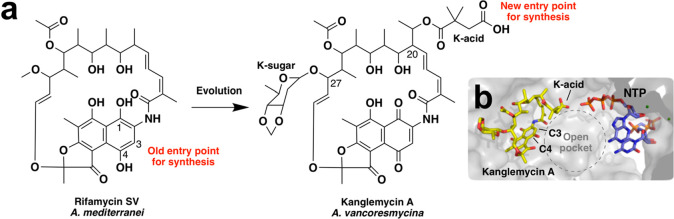
Figure 1.
Structures of rifamycin SV and kanglemycin A. (a) Potential entry points for synthesis. The vast majority of previous synthetic modifications have been made at C-3 and/or C-4 of the rifamycin ring system. Evolution of rifamycin SV has resulted in the addition of features such as the K-acid, which are found in other regions of the molecule that have been largely inaccessible for synthesis. The K-acid provides a new entry point for generating novel semisynthetic derivatives. (b) Position of the K-acid relative to the nascent RNA transcript in the RNAP active site. The nucleotides were modeled into the crystal structure of the Mycobacterium smegmatis RNAP crystal structure in complex with Kang A (PDB ID: 6CCE) by superimposition with the RNAP transcription initiation complex from Thermus thermophilus (PDB ID: 4Q4Z).