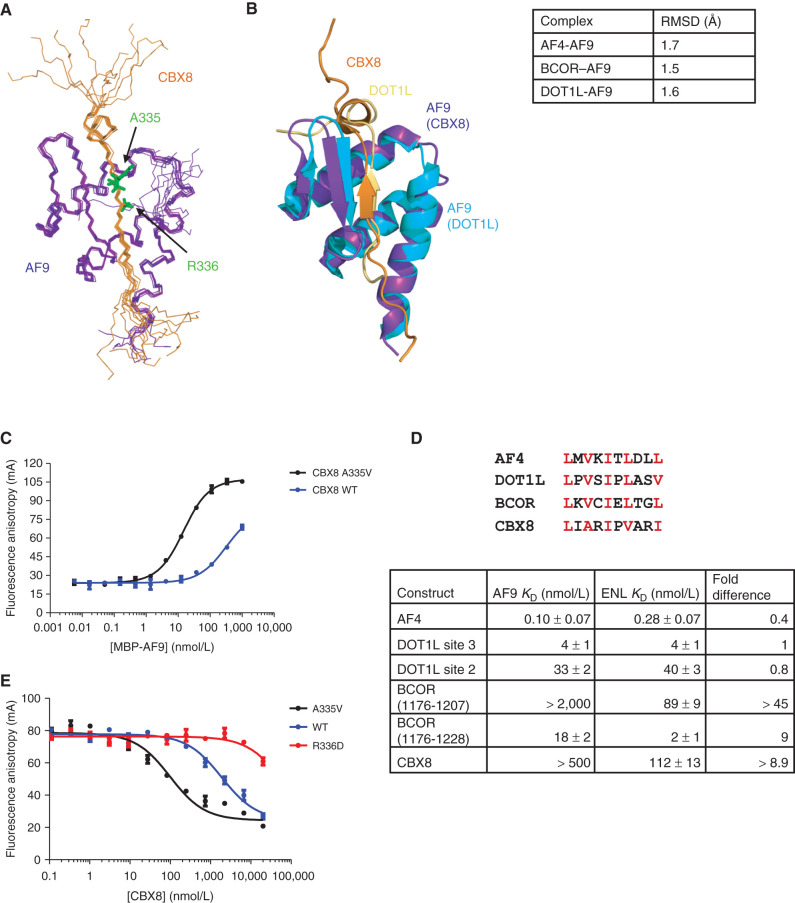
Figure 1.
Structure and binding properties of the CBX8–AF9 AHD complex. A, Ensemble of the 10 lowest energy conformers from the structure calculations with CBX8 in orange and AF9 in purple. B, Alignment of the CBX8–AF9 and DOT1L–AF9 (PDB: 2MV7) structures with CBX8 in orange, its AF9 in purple, DOT1L in gold, and its AF9 in cyan. Backbone RMSD for AF9 residues 502–562 between CBX8–AF9 and the other AF9 complexes is shown in the table on the right. See also Supplementary Table S1 and Supplementary Fig. S1. C, Results of FA assays for binding of MBP-AF9 to fluorescein-labeled CBX8 WT (blue) and A335V (black) peptides. Error bars indicate SEM for three replicates. D, Top, sequence alignment of the AF9 binding motif. Conserved hydrophobic residues are in red. Bottom-comparison of the binding affinities of fluoresceinated peptides for all binding partners for MBP-ENL to affinities for MBP-AF9 measured by FA. Fold change is calculated as AF9 KD/ENL KD. Error bars, SEM for three replicates. E, Results of FA assays for competition of a fluorescein-labeled DOT1L site 3 peptide off of MBP-AF9 by unlabeled CBX8 A335V (black), WT (blue), and R336D (red) peptides. Error bars, SEM for three replicates. For details, see Supplementary Fig. S2.