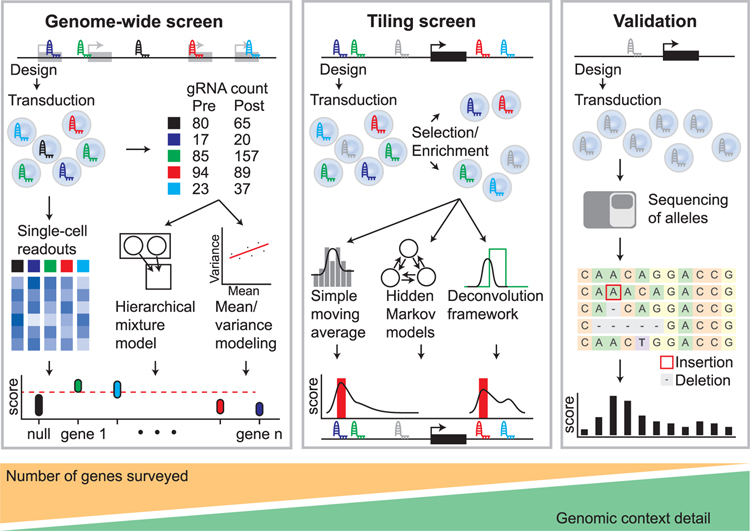
Figure 3.
Overview of Analysis Strategies for Tiling and Gene-Targeted Pooled Screens followed by Screen Validation Genome-wide screening approaches (left panel) utilize gene-targeted gRNA libraries in viral vectors. gRNA abundance is determined before and after phenotypic selection or enrichment. Scores are generated by comparing the relative gRNA abundance in pre-and post-selection populations, and identification of critical genes is performed using mean/variance modeling to address overdispersion or hierarchical mixture models to accommodate gRNA- and gene-specific variation. Alternately, single-cell readouts such as scRNA-seq can be applied to populations of cells to link phenotypes to specific perturbations. Tiling screens (middle panel) are performed by targeting gRNAs across a genomic interval. gRNA abundance is determined before and after phenotypic selection/enrichment. Scores are generated by comparing the relative gRNA abundance in pre-and post-selection populations. The effect of each gRNA can be computed using simple moving averages, hidden Markov models, or deconvolution frameworks. Pooled screen validation (right panel) often involves re-testing gRNAs in an arrayed format in bulk cell populations or individual clones using the techniques for measuring editing at known loci. For example, next-generation sequencing of an individual gRNA target site can be followed by computational analysis to identify generated alleles and calculate a per-base activity score.