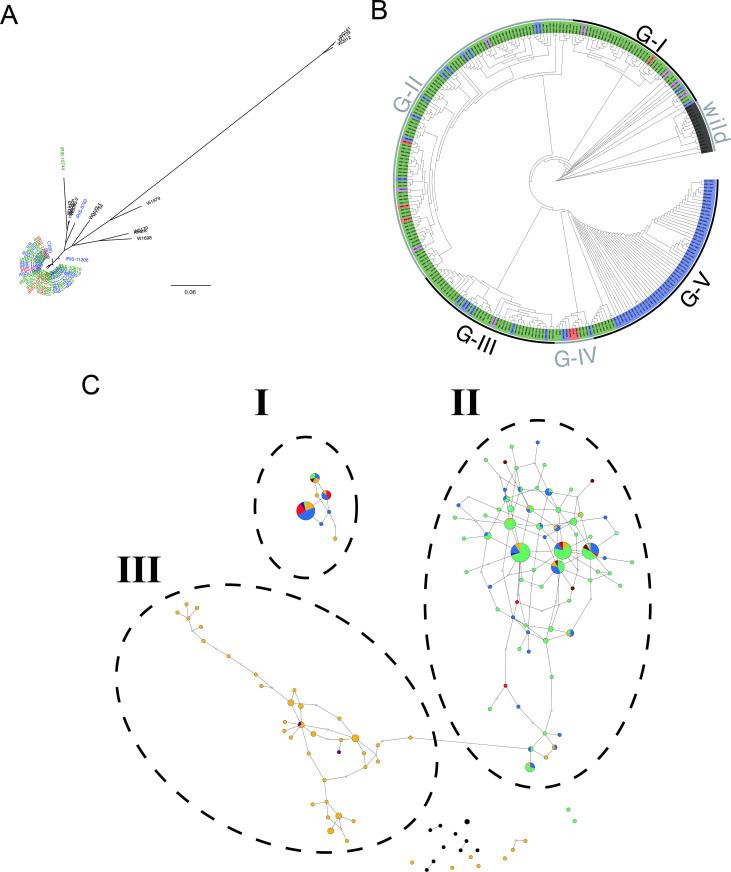
Fig 2. Phylogenic and network analysis of types 7 and 13 Hd1 gene.
Nucleotide changes in the −1000 to + 1000-bp Hd1 region among 3K Asian landraces carrying types 7 and 13 loss-of-function mutation haplotypes as well as 15 Asian wild rice accessions were used. Accession information is in S1 Table. A. Phylogenetic tree of 40 3K accessions carrying type 7 Hd1 mutation and 15 wild rice accessions. The tree was constructed with SNP data by using PHYLIP [44]. Green indicates insular Southeastern Asia areas, red Indochina areas, blue Indian subcontinent, magenta Madagascar and black wild rice species. The bootstrap values determined with 1,000 samples are shown. B. Phylogenetic tree of 262 3K accessions carrying type 13 Hd1 mutation and 15 wild rice accessions. Groups I to V are indicated. Other notes are the same as A. C. Allele network of types 7 and 13 as well as the wild rice accessions was constructed with the TCS program v1.13 [28]. Black circles indicate wild rice accessions, green and blue highlight japonica and indica types from insular Southeastern Asia areas, cyan and red highlight japonica and indica for Indochina areas, purple and orange highlight japonica and indica for Indian subcontinent, and brown highlights indica for Madagascar. Sizes of circles are proportion to the haplotype frequencies. Lines indicate mutation steps.