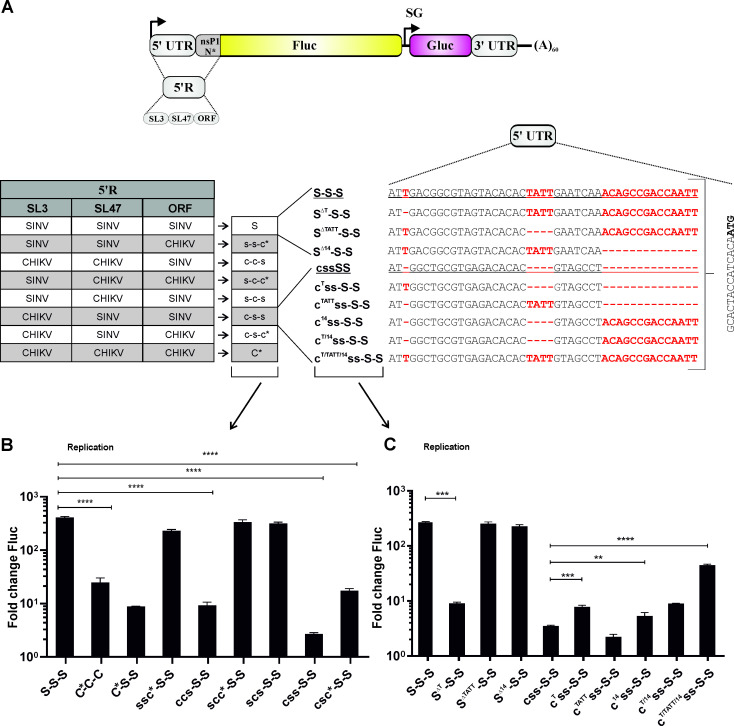
Fig 10. Determinants located at the 5’ end of the template RNA are crucial for SINV replicase.
(A). Schematic representation of template RNAs. Changes introduced into the 5’ region of S-S-S template RNA are shown on left panel. Right panel shows 5’ UTR sequences of modified S-S-S and css-S-S templates. SL structures in CHIKV sequence are designated SL3 and SL47 according to [33]. Other elements and designations are the same as Figs 1A and 9A. (B). U2OS cells grown in 12-well plate were co-transfected with CMV-P1234-SINV and HSPolI-FG-SSS, HSPolI-FG-C*CC, HSPolI-FG-C*SS or derivatives of HSPolI-FG-SSS expressing templates containing indicated swaps in their 5' region. For transfection of control cells CMV-P1234GAA-SINV and HSPolI-FG-SSS were used. (C) U2OS cells grown on 12-well plate were co-transfected with CMV-P1234-SINV and HSPolI-FG-SSS, HSPolI-FG-cssSS or their derivatives expressing templates containing indicated mutations in the SL3 and/or in extreme 5’ end of the template. For transfection of control cells CMV-P1234GAA-SINV was used instead of CMV-P1234-SINV. Cells were lysed 18 h p.t. Fluc activities produced by active replicases were normalized to those measured in control cells. Means + SD of three independent experiments are shown; ** p<0.01, ***p<0.001, ****p<0.0001(Student's unpaired t-test).