Figure 1.
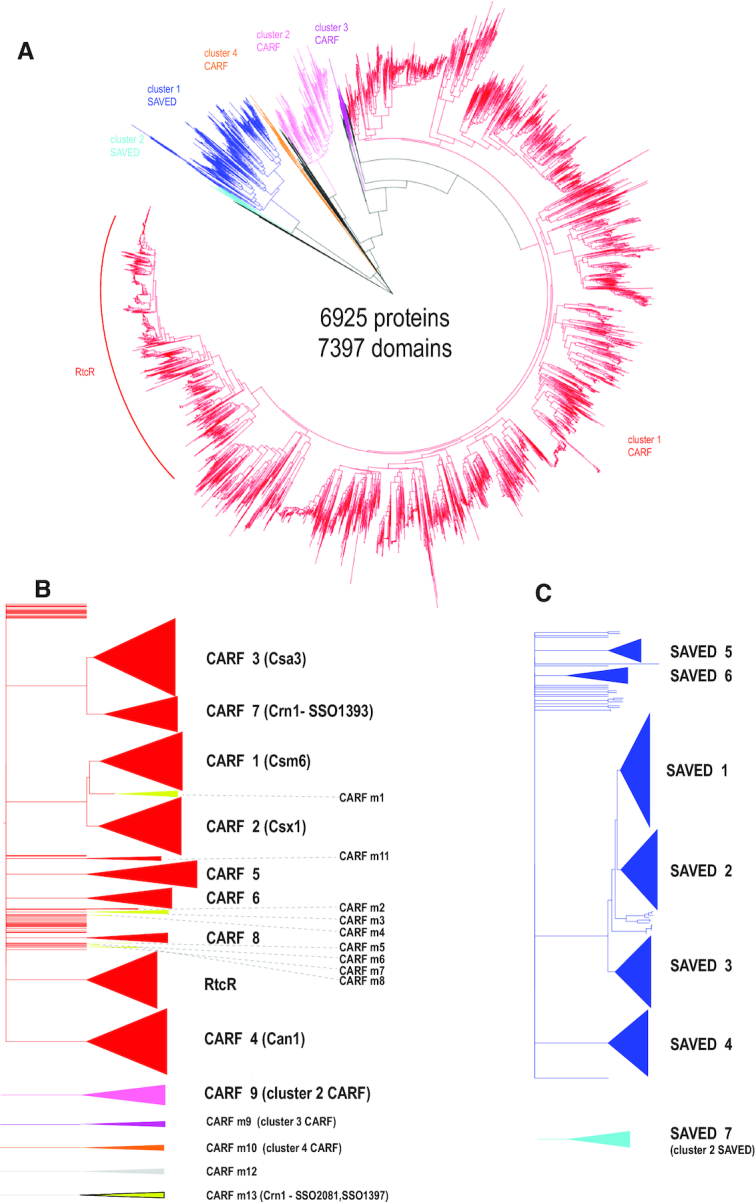
Relationships between CARF and SAVED domain-containing protein sequences. (A) Dendrogram built from the alignment of CARF and SAVED domain sequences only. The dendrogram was built using the ‘hybrid’ approach for sequence classification. Briefly, the FastTree program was used to infer relationships within alignable clusters, and the relationships between these clusters were inferred from HHalign pairwise scores using the matrix-based UPGMA method as described in detail previously (1). Distinct major alignable clusters are color coded. (B) Dendrogram built using alignment of complete amino acid sequences of CARF domain-containing proteins. Major and minor CARF clades corresponding to well-supported branches that include five or more sequences from diverse genomes are shown schematically on the right. The color coding is the same as in panel A. CARF_m13 group sequences are highly divergent and are included only in the second dendrogram. (C) Dendrogram built from the alignment of complete amino acid sequences of SAVED domain-containing proteins. Seven SAVED clades corresponding to well-supported branches that include 5 or more sequences from diverse genomes are shown schematically on the right. The color coding is the same as in panel A. The dendrograms in panels B and C were built using the same approach as the dendrogram in panel A. The subtrees including the sequences from the major cluster CARF1 (red) and SAVED (blue) were extracted from the tree built using complete protein sequences. Common names used in the literature are indicated in parentheses.
