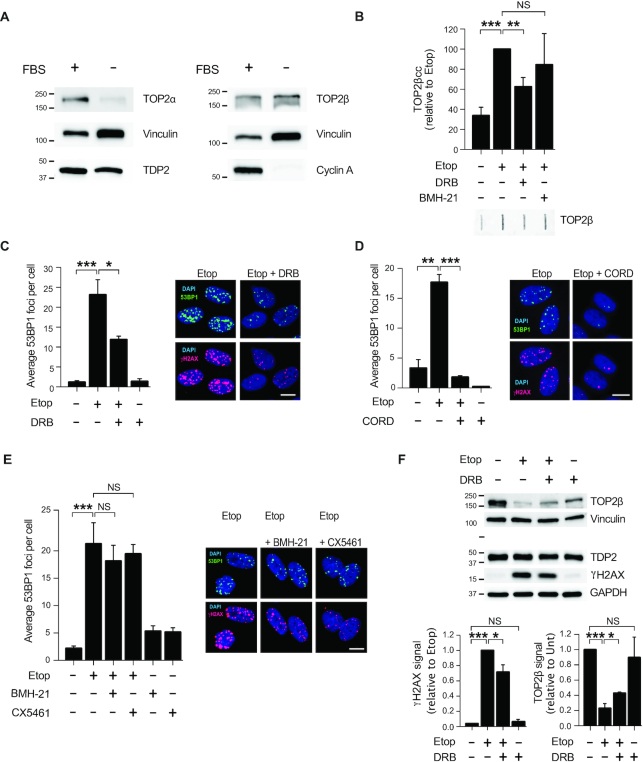
Figure 1.
Transcription by RNAP II promotes TOP2β-induced DSBs in non-cycling cells. (A) Protein blots of the indicated proteins in proliferating (FBS+) and serum-starved (FBS–) RPE-1 cells. Molecular weight markers are in kDa. (B) Analysis of TOP2β cleavage-complexes (TOP2βcc) by ICE assay. Serum-starved RPE-1 cells were treated with 50 μM etoposide for 1 h. Where indicated, cells were pre-incubated with DRB (100 μM) or BMH21 (2 μM) for 1 h prior to etoposide treatment. Data are the mean (± s.e.m.) of five independent experiments. Statistical significance was determined by t-test (**P < 0.01, ***P < 0.001, NS, not significant). (C) 53BP1 foci in serum-starved RPE-1 cells following following treatment with 20 μM etoposide for 1 h. Where indicated, cells were pre-incubated with DRB (100 μM) for 3 h prior to etoposide treatment. Representative images of 53BP1 foci (green), γH2AX foci (red) and DAPI counterstain (blue) are shown. Scale bar, 10 μm. In all cases, data are the mean (± s.e.m.) of at least three independent experiments. Statistical significance was determined by t-test (*P< 0.05, **P< 0.01, ***P< 0.001, NS, not significant). (D) 53BP1 foci in serum-starved RPE-1 cells following etoposide treatment. Where indicated, cells were pre-incubated with 100 μM cordycepin (CORD) for 2 h prior to etoposide treatment. Other details as in (C). (E) 53BP1 foci in serum-starved RPE-1 cells following etoposide treatment. Where indicated, cells were pre-incubated with BMH21 (2 μM) for 3 h or CX5461 (20 μM) for 3 h prior to etoposide treatment. Other details as in (C). (F) Top, protein blots of the indicated proteins in serum-starved RPE-1 cells following treatment with 100 μM etoposide for 6 h. Where indicated, cells were pre-incubated with DRB (100 μM) for 3 h prior to etoposide treatment. Molecular weight markers are in kDa. Bottom, quantification of TOP2β and γH2AX signal normalized with respect to GAPDH. Data are the mean (± s.e.m.) of three independent experiments. Statistical significance was determined by t-test (*P< 0.05, ***P< 0.001, NS, not significant).