Figure 6.
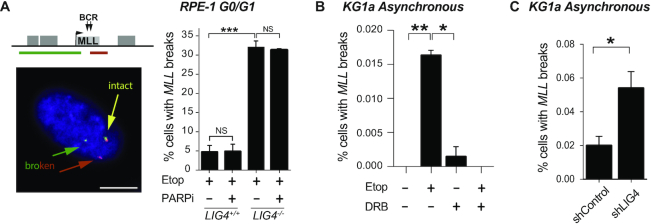
cNHEJ pathway repairs transcription-dependent DSBs at MLL. (A) Detection of broken MLL loci by FISH in serum-starved wild-type and LIG4−/− RPE-1 cells following mock-treatment (DMSO) or treatment with 100 μM etoposide for 6 h. Where indicated, cells were pre-incubated with PARP inhibitor KU58948 (1 μM) for 1 h prior to etoposide treatment. Left, representative images of intact (yellow) and broken (red and green) MLL loci. Scale bar 10 μm. The position of the FISH probes (red and green lines in top cartoon) flanking the MLL breakpoint cluster are shown. Right, data are the mean (± s.e.m.) of at least two independent experiments. Statistical significance was determined by t-test (***P< 0.001, NS, not significant). (B) Detection of broken MLL loci by FISH in KG1a cells following treatment with 100 μM etoposide for 3 h. Where indicated, cells were pre-incubated with DRB (100 μM) for 1 h prior to etoposide treatment. Data are the mean (± s.e.m.) of three independent experiments. Statistical significance was determined by t-test (*P< 0.05, **P< 0.01). (C) Detection of broken MLL loci by FISH in KG1a shRNA LIG4 and control cells following treatment with 100 μM etoposide for 3 h. Data are the mean (± s.e.m.) of three independent experiments. Statistical significance was determined by t-test (*P< 0.05).