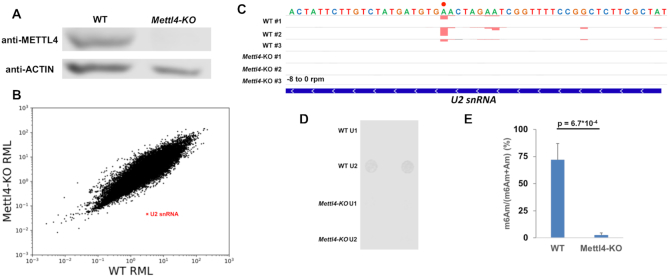
Figure 1.
METTL4 mediates m6Am modification in U2 snRNA. (A) Western blotting of human METTL4 in WT versus Mettl4-KO lysate with ACTIN as a loading control. (B) Scatterplot of average relative methylation level (RML) of sites identified via m6ACE-seq of WT versus Mettl4-KO RNA. U2 snRNA is denoted with a red ‘X’. (C) m6ACE (red) and Input (black) read-start counts (in reads per million mapped or RPM) mapped to U2 snRNA. U2 snRNA m6Am position identified in (B) is denoted by a red dot. Sequence corresponds to the same strand as the m6Am site. Blue horizontal bar represents transcript. (D) Anti-m6A dot blotting of various snRNAs purified from WT versus Mettl4-KO RNA. Duplicate dots are shown. (E) Nucleoside HPLC–MS/MS of m6Am as a percentage of total m6Am and Am in U2 snRNA purified from WT versus Mettl4-KO RNA. Displayed are average and standard deviation error of biological triplicates. One-tailed Student's t-test P-value is shown.