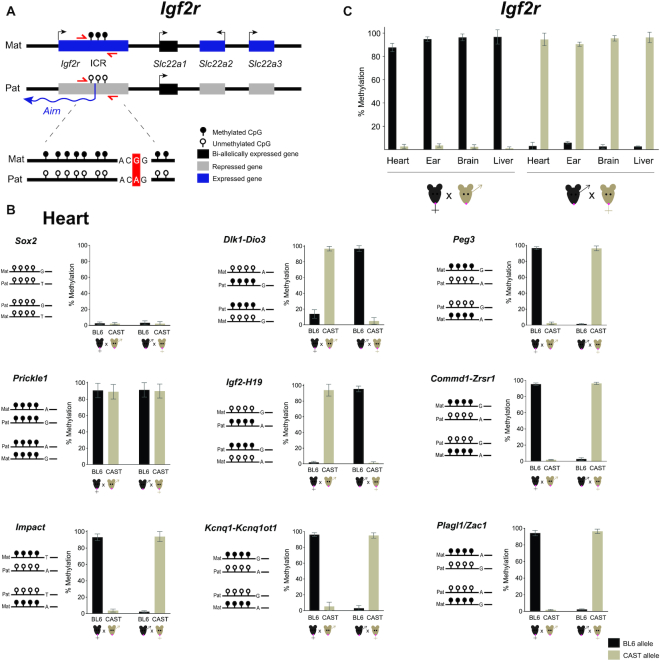
Figure 3.
Allele-specific IMPLICON in F1 adult tissues from C57BL/6J x Cast/EiJ reciprocal crosses. (A) Schematic representation of the Igf2r imprinted cluster; Mat – Maternally inherited chromosome; Pat – Paternally inherited chromosome; ICR – imprinting control region; red arrows – primers to amplify Igf2r ICR; in the scheme below, a single nucleotide polymorphism is highlighted in red; genomic region not drawn to scale. (B) Methylation analysis of Sox2 (unmethylated control), Prickle1 (methylated control) and ICRs of imprinted regions in the heart from F1 hybrid adult mice derived from C57BL/6J × CAST/EiJ reciprocal crosses; Graph represents the mean ± SD methylation levels measured at each CpG within different genomic regions per parental allele in the two F1 hybrid mice; Scheme on the left of each graph represents the expected methylation status of each region (white lollipops – unmethylated CpGs; black lollipops – methylated CpGs; Mat – maternal allele; Pat – paternal allele; black mice (BL6) – C56BL/6J strain; brown mice (CAST) – CAST/EiJ strain; regions are not drawn to scale. (C). Methylation analysis for the Igf2r imprinted cluster in heart, ear, brain and liver of F1 hybrid mice from reciprocal crosses; Graph represents the mean ± SD methylation levels measured for all the CpGs within the Igf2r ICR in each parental allele per mouse.