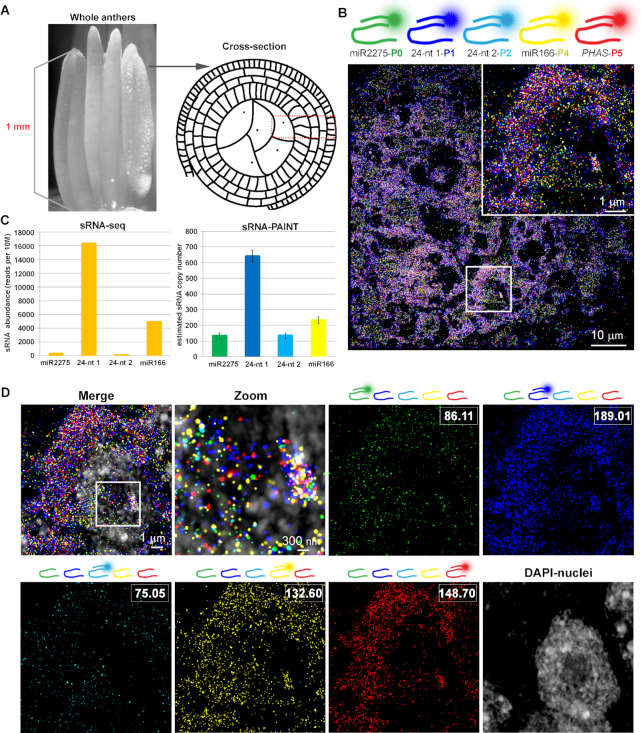
Figure 5.
sRNA-Exchange-PAINT for detection of phasiRNA biogenesis components in premeiotic maize anther. (A) Schematic of maize anther stage and comparison of material used in sRNA-seq and sRNA-PAINT method. (B) Image of sRNA-Exchange-PAINT carried on five targets. Zoomed-in image of the boxed area is shown on the right corner. miR2275 probes were linked with the P0 docking strand and represented with green color. 24-nt phasiRNA 1 probe was linked with P1 docking strand and represented with blue color. 24-nt phasiRNA 2 probe was linked with P2 docking strand and represented with cyan color. miR166 probe was linked with P4 docking strand and represented with yellow color. PHAS lncRNA was linked with P5 docking strand and represented with red color. (C) Comparison of the quantification result of four target miRNAs using sRNA-seq and sRNA-qPAINT. Representative sRNA copy number estimates were calculated by the sum of the binding sites shown in the boxed area using sRNA-qPAINT. (D) Sequential detection of five targets and overlay with the nucleus. Overlay of the channels shows the complexity and density of the small RNAs. Zoomed in image of the boxed area shows single-molecule detection capability with higher-level of multiplexing. The imager strand in each channel is indicated with the diagram above each image, and the quantification of binding sites of each channel is shown in the white box.