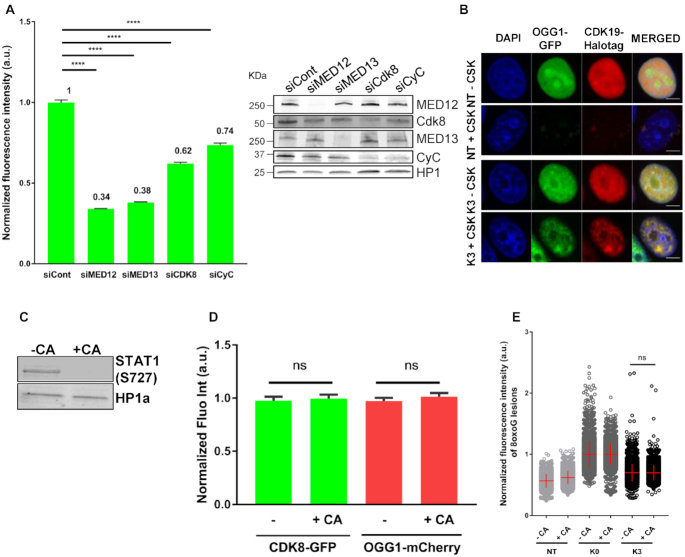
Figure 5.
CDK8 presence but not its enzymatic activity is required for recruitment of OGG1. (A) Quantification of OGG1–GFP fluorescence intensity in non- (NT) and KBrO3- (K3) treated after CSK washing in HeLa OGG1–GFP cells transfected with a siRNA control or siRNAs against the indicated mediator subunits. More than 2000 cells were analysed from two independent experiments. Error bars indicate SEM. Statistical analysis involved a Kruskal–Wallis test. (****) P < 0.0001. Specificity and efficiency of the siRNAs was validated by Western blot. (B) Distribution patterns of OGG1–GFP/CDK19-Halotag in non- (NT) and KBrO3- (K3)-treated cells. Prior to fixation, soluble proteins were removed with CSK when indicated. Scale bar: 5 μm. (C) Efficiency of the CDK8–CDK19 inhibitor CA was evaluated by the phosphorylated state of STAT1 (S727). (D) Quantification of OGG1-mCherry and CDK8-GFP fluorescence intensities in KBrO3 treated cells after CSK washing, in HeLa cells incubated or not with a CDK8 inhibitor (CA). 2900 cells from three independent experiments were analysed. Error bars indicate SEM. Statistical analysis involved a Kruskal–Wallis test. (E) Quantification of 8-oxoG fluorescence intensity in non- (NT) or KBrO3 treated HeLa OGG1–GFP cells fixed just after treatment (K0) or after 3 h of recovery (K3), in the presence or absence of the CDK8/CDK19 inhibitor CA. More than 4000 cells were analysed from three different experiments. The mean of 8-oxoG measured immediately after exposure to KBrO3 (K0H) for each condition was set to 1 and used for normalization. Statistical analysis involved a Kruskal–Wallis test.