Figure 1.
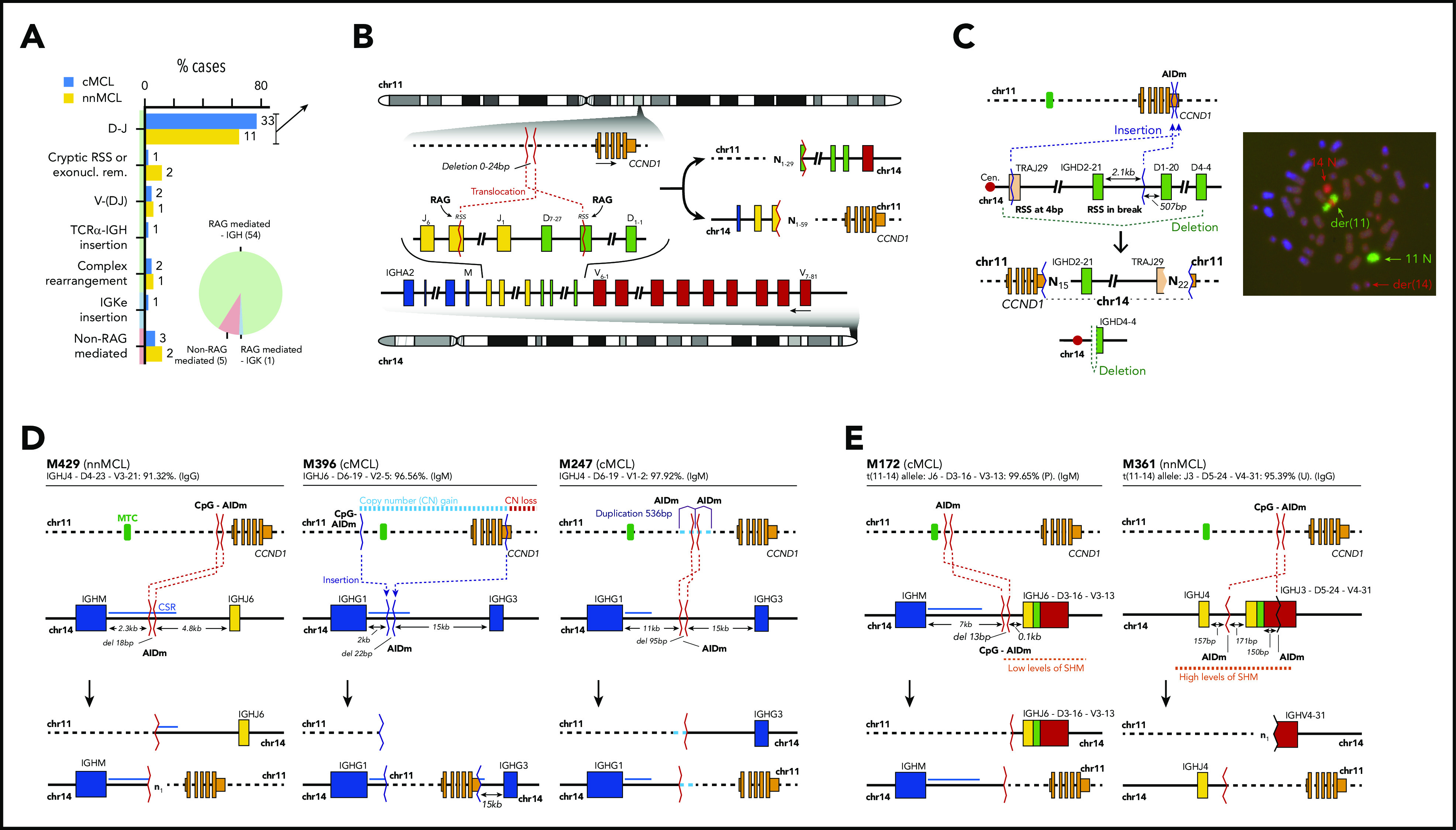
Characterization of the IG breaks of the IG/CCND1 translocation. (A) Distribution of the IG breaks according to their underlying mechanism (RAG or non-RAG mediated) and location of the breakpoints in chr14. (B) Schema of the most recurrent translocation pattern with breaks at IGHD and IGHJ genes, near RSS, likely generated during the first step of an IGH D-J rearrangement. The presence of N-nucleotides in both breakpoints (NX) supports a RAG-mediated process. (C) Unbalanced IG/CCND1 translocation in which virtually the entire 14q arm is inserted at the 3′ UTR of CCND1. In this case, a single event truncated the CCND1 3′ UTR region and placed the IG enhancer near CCND1. The FISH whole-chromosome painting for chr11 and chr14 verifies the ins(11;14)(q13;q11q32) identified by WGS. For the sake of clarity, an interphasic nuclei present in the lower left part of the original picture was masked using Adobe Photoshop. (D) Representation of 3 cases in which the IG breakpoint was likely mediated by AID during CSR. The switch regions 5′ of the IGH constant genes are indicated by a blue line. The productive V(D)J rearrangement, IGHV identity (%), and isotype expression is specified. (E) Representation of 2 cases showing evidence for the involvement of the SHM machinery in the breakpoints of chr14 in already V(D)J-rearranged alleles. P, productive rearrangement; U, unproductive rearrangement.
